Manuscript accepted on : 21-06- 2023
Published online on: 12-07-2023
Plagiarism Check: Yes
Reviewed by: Dr. Karpagam Sundaramurthy
Second Review by: Dr Sabyasachi Banerjee
Final Approval by: Dr. Ahmad Ali
18S rRNA Approach for Identification of Chara L. Species
Rekha Adimulam* and P Sujathamma
and P Sujathamma
Department of Biosciences and Sericulture, Sri Padmavati Mahila Visvavidyalayam (Women’s University), Tirupati-517502, Andhra Pradesh
Corresponding Author E-mail: adimulamrekha@gmail.com
DOI : http://dx.doi.org/10.13005/bbra/3154
ABSTRACT: The study focused on the molecular phylogenetics for the identification of genus Chara species through 18S rRNA genes. Genus Chara sample is collected in the freshwater streams of Talakona region, Chittoor district, Andhra Pradesh and DNA was isolated. The 18S rRNA, gene sequence was used to carry out BLAST with the ‘nr’ database of NCBI GenBank database. First ten sequences were selected based on maximum identity score using multiple alignment software program Clustal W. Ribosomal Database Project was used for generating distance matrix and Molecular Evolutionary Genetics Analysis 6 Software was used for constructing the phylogenetic tree. Based on the maximum identifying score Chara species was identified as Chara foetida.
KEYWORDS: Chara foeitda; Identification; Molecular phylogenetics; Phylogenetic tree
Download this article as:| Copy the following to cite this article: Adimulam R, Sujathamma P. 18S rRNA Approach for Identification of Chara L. Species. Biosci Biotech Res Asia 2023;20(3). |
| Copy the following to cite this URL: Adimulam R, Sujathamma P. 18S rRNA Approach for Identification of Chara L. Species. Biosci Biotech Res Asia 2023;20(3). Available from: https://bit.ly/44I2o34 |
Introduction
Charophyceae is macroscopic algae, habit in both fresh and brackish water, upright stem like structure with branches, in addition to whorl of secondary branches 15. These members are in close relation to higher plants due to the presence of similar pigmentation and reproductive structures 4. They are used as research models in biotechnology, genetic engineering, biochemistry, cell cycle and physiology 5. Algae have the great importance in ecological and economical context. Hence the availability of systematic details, diversity and distribution is essential for future research work. Systematics deals with identification of plant and evolutionary relationships 3. They are studied by the classical and molecular methods. In classical method the plants are identified based on the morphological characteristics and inmolecular method plant are identified with the DNA sequence 1. Identification of organism through molecular phylogenetic study can be done in short gene from a uniform locality on a genome. Each organism has different type of DNA barcode, so it has the great importance in the taxonomy, evolution and phylogenetical arrangement 2. The first step of the Molecular phylogenetic studies is to select the suitable primers, amplified the barcode region PCR. In plant species which show similar morphology, differences can be identified through molecular phylogenetic study 12. Sometimes it is difficult to identify the Chara speciesin which do not bear sex organs 15. In such conditions the Molecular phylogenetic study helps in the identification of Organism. Anyone who has the Basic knowledge on DNA technology is adequate to identify the organism. Molecular phylogenetic study is not the only solution for all taxonomical problems, but it provides the effective nomenclature 10 and identification for the unknown or un recognized species. The organisms which are not identified in the classical methods can be identified by the molecular phylogenetic study.
Material and Methods
Collection and Culture of Chara species
Chara sample was collected from the fresh water streams of Talakona region, Chittoor district, Andhra Pradesh. The sample was isolated from other associated algae, cleaned thoroughly with the water 19.Chara sample was cultured in in vivo condition and named as CH9.
Molecular Phylogenetic
DNA was isolated from the samples with DNeasy plant Mini Kit(Quiagen, Hilden, Germany). 1.0 % agarose gel was used for quality assessment, a high-molecular weight single band DNA has been identified.
CDMFP and CDMRP primers were used for amplifying the fragments of 18S rRNA gene. 700 bp a single discrete PCR amplicon band identified in the Sample when resolved in agarose gel. CDMFP and CDMRP primers are carried out the DNA sequencing reaction. Forward and reverse PCR amplicon was used in BDT v3.1 Cycle (sequencing kit on ABI 3730xl)on Genetic analyzer for eliminating contaminants.
Aligner software used for generating Consensus sequence of 18S rRNA from forward and reverse sequence data. The 18S rRNA gene sequence was used to carry out BLAST with the ‘nr’ database of NCBI GenBank database. Ten sequences are selected based on the Maximum identity score and multiple alignment software program Clustal W was used for alignment. Ribosomal Database Project was used for generating distance matrix. Molecular Evolutionary Genetics Analysis 6 software was used for constructing the phylogenetic tree.
Results
Morphological Characters
Plant is 30 cm long, 620 µm wide and 6-9 whorls branch lets,incurved, terminal cell with 3 projections.
Collection number: Talakona surrounding areas.
Distribution: First report from Andhra Pradesh [6]
 |
Figure 1: Chara foeitda.
|
Molecular Phylogenetic
Charaspecies shows 91.16% homology to the Charafoeitda. So, Chara sample was identified as Chara foeitda
Maximum Likelihood method (Tamura-Neimodel) are used for the evolutionary history 16,17 CH9 sample tree shows highest log likelihood at (-1309.77). 11 nucleotide sequences are involvedin analysis and Codon positions included were 1st+2nd+3rd+Noncoding.In Chara foetida total of 737 positions in the final data set. Evolutionary analyses were conducted in MEGA 6.
 |
Figure 2: Genomic DNA and Amplicon QC data
|
CDMRP Primer
 |
Figure 3: CDMFP Primer
|
Table 1: Sequences producing significant alignment
|
Description |
Max score |
Total score |
Query cover |
E value |
Ident |
Accession |
|
C.foetida |
968 |
968 |
99% |
0 |
91.16% |
X70704.1 |
|
Chara andina |
965 |
965 |
99% |
0 |
91.02% |
AF032724.1 |
|
Chara globularis voucher NY:02282230 |
963 |
963 |
99% |
0 |
91.02% |
KR080214.1 |
|
Chara braunii voucher NY:02282229 |
963 |
963 |
99% |
0 |
91.02% |
KR080213.1 |
|
Chara intermedia , isolate TK31 |
963 |
963 |
99% |
0 |
91.02% |
HF913651.2 |
|
Chara globularis , isolate GJ29 |
963 |
963 |
99% |
0 |
91.02% |
HF913643.2 |
|
Chara aspera , isolate BR02 |
963 |
963 |
99% |
0 |
91.02% |
HF913640.2 |
|
Chara australis strain X-067 |
963 |
963 |
99% |
0 |
91.02% |
AY823707.1 |
|
Chara globularis |
963 |
963 |
99% |
0 |
91.02% |
Y16465.1 |
|
Chara polyacantha |
963 |
963 |
99% |
0 |
91.02% |
AF032742.1 |
 |
Figure 4: Sanger Seq Chromatogram data file.
|
Table 2: Distance matrix
|
|
1 |
2 |
3 |
4 |
5 |
6 |
7 |
8 |
9 |
10 |
|
1.CH9 |
|
|
|
|
|
|
|
|
|
|
|
2.X70704.1 |
0.075 |
|
|
|
|
|
|
|
|
|
|
3.AF032724.1 |
0.075 |
0.003 |
|
|
|
|
|
|
|
|
|
4. KR080214.1 |
0.077 |
0.004 |
0.004 |
|
|
|
|
|
|
|
|
5.KR080213.1 |
0.080 |
0.007 |
0.004 |
0.009 |
|
|
|
|
|
|
|
6.HF913651.2 |
0.078 |
0.003 |
0.003 |
0.004 |
0.007 |
|
|
|
|
|
|
7.HF913651.2 |
0.077 |
0.004 |
0.004 |
0.000 |
0.009 |
0.0004 |
|
|
|
|
|
8.HF913640.2 |
0.077 |
0.001 |
0.001 |
0.003 |
0.006 |
0.001 |
0.003 |
|
|
|
|
9.AY823707.1 |
0.079 |
0.009 |
0.006 |
0.007 |
0.004 |
0.009 |
0.007 |
0.007 |
|
|
|
10.Y16465.1 |
0.077 |
0.004 |
0.004 |
0.000 |
0.009 |
0.004 |
0.000 |
0.003 |
0.007 |
|
|
11.AF032742.1 |
0.077 |
0.001 |
0.001 |
0.003 |
0.006 |
0.001 |
0.003 |
0.000 |
0.007 |
0.003 |
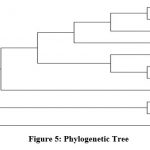 |
Figure 4: Phylogenetic Tree
|
Discussion
The taxonomical studies based on the morphology sometimes may leads to the misidentification 11. To overcome to this, molecular approaches are used. The 18s RNA, matK, ITS2 and rbcl genes are most widely used and effective methods in Molecular phylogenetic 13.
The class Charophyceae members are mostly benthic in nature, complex structure. The identification of these members is difficult due to the presence of phenotypic plasticity. The present study identified the macroscopic algae Chara with molecular phylogenetic technique. Chara species is identified by sequencing of 18s RNA gene analysis, namely Chara foeitda. The sample code CH9 shows 91.16% similarity to the Chara foeitda.
Monique Turmelet al (2006) studied the atpB, rbcl, 18S rRNA and mitochondria (nad5) of Chara vulgaris and it showed closest relation to the green plants of land 18. Hidetoshi identified the Chara globularis with molecular phylogenetic based on the rbcl gene sequencing 7. Schneider et al identified the 14 species of Chara with barcodes like ITS2, Mat K and rbcl genes from Europe 14. Jacek& Michal studied the Chara globularis Var. tenuispina and Chara globularis phylogenetic relationship 8. Based on the atpB, matK and rbcl sequences and identified C. tenuispina distinct species from Chara globularis 9. The present study is one of the pioneer attempt to the molecular phylogenetic for Charophyceae of India.
Conclusion
Chara species are identified as Chara foetida based on the molecular phylogenetic. For the identification 18S rRNA sequence was used to carry out BLAST with the ‘nr’ database of NCBI GenBank database. First ten sequences were selected based on maximum identity score using multiple alignment software program Clustal W. Ribosomal Database Project was used for generating distance matrix and Molecular Evolutionary Genetics Analysis 6 Software was used for constructing the phylogenetic tree. Based on the maximum identifying score Chara species was identified as Chara foetida.
Conflict of Interest
The authors declare that there is no conflict of interest regarding the publication of this paper.
References
- Akash P, Bakhtiyar AS, Bharat G, Pankaj P, Beena P, Rbcl Marker based approach for molecular identification of Arthrospira and Dunaliella isolates from non- Axenic culture, Journal of Genetics and Genetic Engineering, 2018, 2:24-34. https://www.researchgate.net/publication/328118071_ Rbcl_Marker_Based_Approach_for_Molecular_Identification_of_Arthrospira_and_Dunaliella_Isolates_ from_Non-Axenic_Cultures
- Alberts B, Johnson A, Lewis J, Molecular Biology of the Cell. Garland Science; Studying Gene Expression and Function, Garland Science, New York, 2002, 1-50, https://www.ncbi.nlm.nih.gov/ books/NBK26818/
- Braun .A, Esquissemono graphique du genre Chara. Ann. Sci. Nat., sér., 1834, 1: 349-357.
- David. D, Iben.S, Zoe .P, Charophytes: Evolutionary Ancestors of plants and Emerging models for plant research, Frontiers, USA, 2017, 1-5, doi: 10.3389/fpls.2017.00338
CrossRef - David. D, Zoe.P, Iben.S, Charophytes: Evolutinorygaints and emerging model organisam, Frontiers, USA, 2016, 6-10, https://doi.org/10.3389/fpls.2016.01470
CrossRef - Gupta RK, Algae of India Vol.2. Achecklist of Chlorophyceae, Xanthophyceae, Chryophyceae and Euglenophyceae, Botanical survey of India, Kolkata, 2012, 9-331.
- Hidetoshi Sakayama, FumieKasai, Hisayoshi Nozaki, Makoto M. Watanabe, Masanobu Kawachi, Mikao Shigyo, Jun Nishihiro, Izumi Washitani, Lothar Krienitz and Motomi Ito, Taxonamic re-examination of Chara globularis (Charales, Charophyceae) from Japan based on oospore morphology and gene sequences and their description of c. Leptospora Sp.Nov, Journal of Phycology, 2009, 45: 917-927, https://doi.org/10.1111/j.1529-8817.2009.00700
CrossRef - Jacek U, Hidetoshi S, Taxonomical analysis of closely related species of CharaL.section Hartmania (Streptophyta: Charles ) based on morphological and molecular data, Fottea, 2017A, 17:222-239, DOI:10.5507/fot.2017.004
CrossRef - Jacek U, Michal C, Taxonomical status of Chratenupina A. Br. Based on LM morphology, matK, atpB, and rbcl of cpDNA sequences, Fottea, 2017B, 17:20-33, DOI: 10.5507/fot.2016.011
CrossRef - Jahn C.E., Amy O. C, David K. W. Evaluation of isolation methods and RNA integrity for bacterial RNA quantitation, Journal of Microbiological Methods, 2008, 75:318–324, doi.org/10.1016/ j.mimet.2008.07.004.
CrossRef - Karan T, Erenler R, Altunar Z, Isolation and Molecular identification of some blue green algae (cyanobacteria) from Fresh water sites in Tokat Province Turkey, Turkish Journal of Agriculture – Food Science and Technology, 2017, 5:1371-1378, doi.org./10.24925/turjaf.v5i11.1371-1378.1470.
CrossRef - Lorenz TC, Polymerase chain reaction: basic protocol plus trouble shooting and optimization strategies, Journal of visualized experiments 2012, 63, doi:10.3791/3998.
CrossRef - Monique T, Christian O, Claude L The Chloroplast genome sequence of Chara vulgaris Sheds new light into the closet green algal relatives of land plants, Molecular biology and evolution, 2006 , 23: 1324-1338, doi:10.1093/molbev/msk018.
CrossRef - Prakash S, Sanjay K.V, Identification and characterization of green microalgae, Scenedesmus Sp. and Auctodesmus obliquus MCC33 isolated from Industrial polluted site using morphological and molecular markers, International Journal of Applied Sciences and Biotechnology, 2018, 5: 415-422, doi.10.3126/ijasbt.v5i4.18083.
CrossRef - Schneider SC, Susanne, Rodrigues A, Anuar R, Therese MTF, Ballot A, NDNA barcoding the genus Chara: Molecular evidence recovers fewer taxa than the classical morphology approach, Journal of Phycology, 2015, 51: 367-380, doi: 10.1111/jpy.12282.
CrossRef - Subramanian, Indian Charophyta, MJP Publishers, Chennai, 2018, 45- 168.
- Tamura K., Nei M, Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Molecular Biology and Evolution, 1993, 10:512-526, doi: 10.1093/oxfordjournals.molbev.a040023.
CrossRef - Tamura K., Glen S, Daniel P, Alan F, Sudhir K, MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Molecular Biology and Evolution, 2013, 30: 2725-2729, doi: 10.1093/molbev/mst197.
CrossRef - Zulkarnain C Fadjria, N., Armaini, Armaini, Zainul, Rahadian, Isolation and molecular identification of Fresh water microalgae in Maninjau lake West Sumatra, Der Pharmacia Lettre, 2016 , 8:177-187, https://www.researchgate.net/publication/316888807_Isolation_and_molecular_identification_of_freshwater_microalgae_in_Maninjau_Lake_West_Sumatra.

This work is licensed under a Creative Commons Attribution 4.0 International License.





