Manuscript accepted on : 08-03-2022
Published online on: 24-03-2022
Plagiarism Check: Yes
Reviewed by: Dr. Robert Susło
Second Review by: Dr. Ana Cláudia Coelho
Final Approval by: Dr. Imran Ali
Escherichia coli Strains in Patients with Inflammatory Bowel Diseases; A review
Hadba Al-Amrah* Hanan Alotaibi
Hanan Alotaibi  and Nemat Sadiq
and Nemat Sadiq
Department of Biological Sciences, Faculty of Science, King Abdulaziz University, Jeddah-21589, Saudi Arabia
Corresponding Author E-mail: hggaber@kau.edu.sa
DOI : http://dx.doi.org/10.13005/bbra/2966
ABSTRACT:
Gastrointestinal tract conditions, including inflammatory bowel diseases (IBDs) such as ulcerative colitis (UC) and Crohn’s disease, have been linked to adhesive invasive Escherichia coli (AIEC) pathotypes, with comparable pathogenic properties, although the incidence of AIEC with UC and CD is generally undetermined. While a significant advance has been made in understanding the pathogenic processes of AIEC since it was first characterized a decade ago, the molecular basis that determines the phenotypic features of AIEC pathotypes is still unknown. This article reviews studies that examine the prevalence of E. coli in patients with IBD and discusses its pathophysiological role.
KEYWORDS: Adherent Invasive Escherichia Coli; Crohn’s Disease; Epidemiology; Inflammatory Bowel Disease; Pathogenesis
Download this article as:| Copy the following to cite this article: Al-Amrah H, Alotaibi H, Sadiq N. Escherichia coli Strains in Patients with Inflammatory Bowel Diseases; A review. Biosci Biotech Res Asia 2022;19(1). |
| Copy the following to cite this URL: Al-Amrah H, Alotaibi H, Sadiq N. Escherichia coli Strains in Patients with Inflammatory Bowel Diseases; A review. Biosci Biotech Res Asia 2022;19(1).Available from: https://bit.ly/3LeweCK |
Introduction
Inflammatory bowel disease (IBDs, most predominantly Crohn’s disease (CD) and ulcerative colitis (UC) 1, 2 subtypes, are intestinal conditions that involve persistent inflammation of the gastrointestinal tract 3. IBD is a severe chronic inflammatory illness of the intestine affecting more than 0.3 % of people, with UC 4 and CD 5, 6 being the most common. Internationally, IBD is more common in wealthy countries although is becoming increasingly prevalent in developing countries7. Although the cause of IBD is not fully determined, a growing body of research suggests morbidity is strongly linked to hereditary susceptibility. Additional variables, such as nutrition, tissue damage linked with immune system disturbances, and aberrant gut microbiota, may be implicated, as evidenced by mouse models of IBD. It is important to note that the genetic and microbiota-related origin of IBD may be connected. Recently, scientists may have made a fascinating discovery that points to a probable cause of CD: individuals with CARD15/NOD gene mutations, which rely on lower nuclear factor kappa B activation (NF-𝜅B), proinflammatory cytokine production, and defensin secretion, are prone to CD development 8, 9.
Inflammatory Bowel Disease and the Intestinal Bacteriome
IBDs have confounded immunologists and gastroenterologists since their first description between 75 and 100 years ago. Currently, novel investigative approaches are rapidly leading to improved knowledge of key pathophysiologic mechanisms connected to these disorders, paving the way for effective therapies 10. Patients with UC report abdominal discomfort, diarrhea, weight loss, rectal bleeding, fever, and exhaustion as gastrointestinal and systemic symptoms. Patients with CD patients can evolve intestinal strictures and fistulae among parts of the intestinal and between the colon, skin, and other organs. The symptoms of UC are virtually identical to those of CD excepting no formation of fistula with UC. Both UC and CD are usually chronic and relapsing 11.
In humans, the gut microbiota composition includes about 1000 bacteria species, five Archaea genera, 66 fungus genera, and an ill-defined number of viruses, predominantly bacteriophages. These are essential for the immune system and other physiological systems which cannot function without this intricate ecosystem. However, the gut microbiota does not initiate intestinal inflammation independently, rather, abnormalities in the microbiota (i.e., dysbiosis) or the presence of commensal bacteria, with higher virulence in IBD patients, may trigger an overactive anti-microbial immune response 12. CD involves an extreme interleukin (IL-12/IL-23) and interferon-gamma/interleukin-17 (IFN-γ/IL-17) production in the small bowel and full-thickness intestinal wall inflammation and discontinuous colon ulceration, which frequently includes granulomas. By contrast, UC is linked to excessive IL-13 production and principally influences the colon, with a continued mucosa inflammation involving the rectum and extending proximally 11 (Fig.1).While strongly correlative in terms of presentation and symptoms, both CD and UC have distinct clinical characteristics. There are various serums that differentiate the primary types of IBD and reflect their activity or treatment. Furthermore, an evaluation of anti-microbial antibodies and T cell response to commensal gut bacteriome prevalent in healthy people found significant differences with CD patients13. These are important distinctions between CD and UC that suggest that angiogenesis and inflammation regulation may differ. Thus, IBDs occur in immunocompromised patients (such as chronic granulomatous illness and variable immunodeficiency) with hereditary conditions (like Hermansky-Pudlak syndrome), meaning various immune components have a role in IBD susceptibility. Many researchers examining the pathogenesis of IBD believe that it is caused by an interaction between the gut’s bacterial microflora and the mucosal immune system 14. Thus, the bacterial microflora plays an essential role in the etiology of IBDs; if the microflora is quantitatively and qualitatively normal, the disease defect is found in the mucosal immune system 15. The normal state of immunologic tolerance to microbial antigens in the gut is disrupted in IBD cases, either by the presence of a defective mucosal effector T cell population that overreacts to usual bacterial antigens or by the presence of a defective mucosal T-regulatory cell population that under-reacts to normal microorganism antigens 10. An alternative cause is that the gut microflora has a fundamental abnormality in the abundance or kind of microbiota that make up the community, or the degree to which the organisms interact with the mucosal immune system is abnormal. Despite the fact that numerous microorganisms have been explored as causative factors in the aetiopathogenesis of IBD, the disease can lead to a loss of tolerance since the microbiota is able to drive a normal immune system to respond excessively to microbial antigens. Mycobacterium paratuberculosis, Listeria monocytogenes, Chlamydia pneumonia, Escherichia coli (E. coli), and other bacteria are among these pathogens. The dynamic equilibrium between intestinal bacteria, particularly commensal flora, and the host defense systems at the intestinal mucosa, as well as their role at the outset and connection with intestinal inflammation, are currently receiving greater attention 16. Also, changes in the gut bacterial flora caused by environmental, and specifically dietary, factors are thought to have a significant role in IBD etiology17.
Differences in Gut Microbiota in Ibd
IBD is an autoimmune condition that necessitates the presence of commensal bacteria in the gut. Many studies have propelled the idea that IBD infections are caused by an overactive immune response to a normal member of the gut microbiota18. Duchmann et al. found cells derived from gut IBD tissue when cultured with sonicates of autologous or heterologous intestinal microbes displayed stimulation; by contrast, the cells from the control respond to sonicates of heterologous microbiota only 19. The most widely sketched pathogenesis of IBD is that the sickness is largely a result of the abundance of specific microbiota which triggers a pathological immunological response in the mucosal immune system20. There are two bodies of evidence 3 that support the autoimmune response theory. The first suggests that IBD is linked to pathogenic organisms that cause a low-grade infection of the mucous and as a result elicit an inflammatory response. The second shows that patients with IBD have a defective epithelial barrier that allows nonpathogenic organisms to increase close to parts of the mucosal immune system, provoking inflammatory response3. To asses the first body of evidence, we can examine the findings of recent studies on microbiota linked with the mucosa (rather than the stool microbiota), which are more likely to give rise to infection. A number of studies have found that biopsy tissue from patients with IBD had greater levels of mucosa-linked bacteria in the mucus layer and on the epithelial tissues than tissue of healthy people 3. In IBD research, many studies have found an increased abundance of mucosa-associated microbiota(21-23). In one case, a pathogen-like invasive E. coli was found in the mucosa of 20 % and 40 % of ileal mucosa samples from patients with CD compared to 6 % found in the mucosa of samples from healthy individuals. Also, invasive bacteria were found in roughly 4% of colonic samples from control patients and CD patients compared to 12 % of patients with UC samples (24). A study by Martin et al., however, has cast doubt on the significance of such findings, reporting that the majority of patients with CD and a significant number of healthy people, have substantially higher rates of mucosa-adherent microorganisms (80 % in CD patients and 40 % in healthy individuals)(24). Dietary types and environmental upset that would ordinarily affect species structure and short chain fatty acid (SCFA) levels frequently have a significant effect on the gut microbiota. IBD is marked by long-term alterations in the gut microflora, which are linked to inflammation in the intestine. A substantial drop of butyrate-producing obligate anaerobes from the Firmicutes phylum, the most abundant of which is Faecalibacterium prausnitzii, and is a typical biomarker of IBD(25, 26). Since dysbiosis of obligate and facultative anaerobic bacteria characterize IBD imbalance, it has been argued that oxygen and reactive oxygen species have an essential role in its pathogenesis. Indeed, according to the ‘oxygen hypothesis’, prolonged intestinal inflammation causes an increase in the liberation of oxygen-carrying hemoglobin and reactive oxygen species within the lumen, resulting in a milieu that promotes facultative anaerobic microorganisms. Increased inflammation results from the decrease of obligate anaerobes like F. Prausnitzii that release anti-inflammatory chemicals, causing a feedback loop that exacerbates the disease process 16, 27 (Fig. 1).
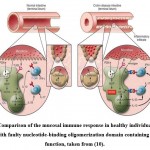 |
Figure 1: Comparison of the mucosal immune response in healthy individuals and CD patients with faulty nucleotide-binding oligomerization domain containing 2 (NOD2) function, taken from (10). |
The toll-like receptor 2 (TLR2) on the surface of dendritic cells in the gut lamina propria detects peptidoglycan (PGN) produced on commensal bacteria walls (left inset, Fig. 1). sensed by TLR2, and as a result, there is NF-𝜅B downstream activation, which is a critical transcription factor essential for the recognition of cells that produce IFN-γ and IL-17, the pro inflammatory cytokines that are thought to cause CD. PGN, by contrast, is broken down in endosomes and so serves as a source of muramyl dipeptide (MDP), a molecule that nucleotide-binding oligomerization domain containing 2(NOD2) detects and activates. As a result of this activation, PGN-mediated NF-𝜅B activation is inhibited, resulting in down-regulation of TLR-induced cytokine production. Since NOD2 modulation is disrupted in individuals with CD that have NOD2 mutations (right inset, Fig.1), the innate immune ‘thermostat’ of the gut is set at a greater level of proinflammatory cytokine production. Inflammation and disease stem from this and T cell response to mucosal antigens. Increased bacterial abundance in the terminal ileum’s crypts results in greater activation of a mucosal immune system already functioning at a higher grade 10.
E. Coli Pathgenicity
Within a few hours of birth, E. coli colonizes the gut system of newborns. E. coli and its mammalian host colon co-exist in harmony, known as mutualism. It is a strong rival in the gut microflora against anaerobes and facultative anaerobes; it is excluded in immunocompromised hosts or when the usual gut barriers crossed, although E. coli seldom cause illness. Escherich, however, has claimed that specific E. coli might be linked with disease, indicating which E. coli are implicated in infections of the colon and urinary system. Through DNA horizontal transfer of transposons, bacteriophages, and plasmids, particular E. coli have gained unique virulence factors. These improve the capacity of this bacteria to fit into new habitats, allowing them to cause a wider range of illnesses. There are six gastrointestinal tract pathogenic E. coli (IPEC) bacteria linked to GI tract diseases in humans: enteroinvasive E. coli (EIEC), enteropathogenic E. coli (EPEC), enterotoxigenic E. coli (ETEC), enteroaggregative E. coli (EAEC), diffusely adherent E. coli (DAEC)), and enterohaemorrhagic E. coli (EHEC)2 (Fig.2). These pathogenic E. coli cause illness via affecting a vast extent of key host cell activities, such as protein synthesis, apoptosis, signal transduction, mitochondrial role, ion secretion, transcription, cytoskeletal role, and cell split. Moreover, regarding toxins and effectors of microorganisms that impact eukaryote processes, all these pathogens must express a variety of fitness and colonization factors that permit the microorganisms to cling to host cells 28. Several studies based on various techniques have found that E. coli is increased in patients with IBD16, 29, 30 (Table 1)
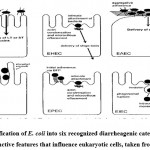 |
Figure 2: Classification of E. coli into six recognized diarrheagenic categories by virtue of distinctive features that influence eukaryotic cells, taken from (31). |
Table 1: Studies on changes in gut E. coli quantity in IBD patients.
| Ref. | Type of sample | Method | Abundance of E. coli in
CD and UC |
| Clapp et al., 2017 (16) | Mucosa | quantitative Polymerase Chain Reaction (qPCR) | Increase in E. coli abundance in CD patients |
| Lopez-Siles et al., 2014 (29)(27) | Mucosa | qPCR | Increase in E. coli abundance in CD patients |
| Sha et al.,
2013 (30) |
Feces | qPCR | Increase in E. coli abundance in CD and UC patients |
| Schwiertz et al.,
2010 (23) |
Feces | qPCR | Increase in E. coli abundance in CD and UC patients |
| Rehman et al.,
2010 (32) |
Mucosa | cloning | High abundance of E. coli in CD and UC patients |
| Martinez-
Medina et al., 2009 (33) |
Biopsies and mucosa | qPCR | High abundance of E. coli a in CD patients |
| Baumgart et al., 2007 (3) | Mucosa | FISH 16S rRNA clone libraries, and qPCR | Increase of E. coli strains in CD patients |
| Mylonaki et al.,
2005 (34) |
Biopsies | FISH | High abundance of AIEC in CD and UC patients |
Adhesive Characteristics of Ibd-Linked E. Coli
It has been found that gut colonization by E. coli is linked to bacteriome adherence by CD-linked E. coli to digestive tract mucosa (to the epithelial cells). The initial stage in the pathogenicity of many organisms with gastrointestinal infections is bacterial adherence to intestinal epithelial cells. The action of the bacteria to colonize the epithelial cells and combat mechanical sweep from the gut is achieved by adhesion. The broad conclusion across a number of studies is that IBD-related adhesion to E. coli can attach to several human cells. According to one study 12, sticky E. coli was identified in 62 % of CD patients and 68 % of UC patients, but only 6 % of healthy control12. In another study, 86 % of E. coli isolated from patients with IBD were sticky, compared to 27 % of E. coli isolated from patients with infective diarrhea and healthy controls 12. Recently, according to Kotlowski et al., 35, E. coli with adhesion factors in CD and UC patient tissues were more abundant than the same in healthy controls. When E. coli bacteria was recovered from the ileum of patients with CD and also from healthy people, it was found that nearly 80 % of E. coli correlating with the ileal mucosa of patients with CD were adhesive compared to 30 % separated from healthy individuals 16. CD-linked E. coli attached to distinguish Caco-2 cells, preferably a mature gut cell sampleb 16. This is in line with the discovery that crypt epithelial cells related to immature cells are seldom implicated in patients with early lesions in connection with CD.
Invasive Characteristics of Ibd-Linked E. Coli.
The lesions that form in CD exist in Peyer’s patches, as seen in early entero-invasive microorganisms, such as Shigella and Salmonella, which can induce GI tract lesions. This supports the notion of an invasive pathogen that initiates CD. Indeed, the aphthous ulcer, the necrosis of microfold cells (M-cells) of Peyer’s lymphoid follicles, is identified as the earliest lesion of CD 36. Invasiveness shigellosis, salmonellosis, yersinia entero, and colitis are primary associated virulence factors of the disease. Many studies have reported the existence of intramucosal E. coli in patients with IBD or mucosa-associated E. coli with invasive traits. Invasive microbiota exists in 29-36 % of patients with CD, in 12-19 % of UC patients, and 3-9 % of healthy individuals 37. The invasive process of LF82 strains separated from a lesion of a CD patient has been extensively studied. LF82 strains efficiently pervade many human epithelial cells, including HEp-2 cells, HCT-8 cells, and the gut cell lines intestine-407 and Caco-2 38. The most invasive bacteria, Yersinia enterocolitica, Shigella Flexner, Listeria monocytogenes, and EIEC, are actin microfilament, but not microtubule-dependent. The uptake of the invasive E. coli and LF82 separated from patients with CD is based on the role of host cell microtubules and actin microfilaments 39. A micropinocytosis-like method of entry was discovered in LF82-infected epithelial cells, distinguished by elongation of the membrane extensions that encircled the bacteria at the areas of contact among the epithelial cells and the entering bacteria. The LF82 strain survives and multiplies in the host cell cytoplasm after lysing the endocytic vacuole. The invasive mechanism of LF82 is unique in that it lacks any of the renowned genetic invasive determinants seen in entering invasive, Shigella strains enteropathogenic E. coli and enterotoxigenic E. coli 38. Also, Pseudomonas aeruginosa and Helicobacter pylori outer membrane vesicles are shown to construct proinflammatory responses, and TLR5, when it reacts with bacterial flagellin, can activate an innate immune reaction 40.
Replication of Cd-Correlating E. Coli in Microphages
Intracellular infections have evolved to combat phagocytosis and survived interior macrophages. The macrophage activation and engagement in persistent antigenic stimulation cells have been the center of the hunt for pathogenic organisms that may prompt CD. Invasive E. coli obtained from CD may live and multiply in a large vacuole of murine macrophages 16. CD-linked invasive E. coli behaves differently in macrophages than other invasive microbiota. While most members of invasive bacteria cause cell death in macrophages that are infected 41, macrophages infected with CD-linked invasive E. coli show no necrosis or apoptosis, even after 24 hours 16. Moreover, CD is related to invasive E. coli that are picked up within macrophages by phagosomes that develop without diverging from the conventional endocytic route and that engage with phagolysosomes. By contrast, many pathogens infiltrate autophagy or escape the normal endocytic process. Microbiota have evolved mechanisms where acidity is a critical sign for triggering the expression of malignancy genes and increase in the severe environment found within these compartments, including cathepsin D proteolytic activity and acid pH 16. Tumor necrosis factor (TNF) is released in substantial levels by macrophages infected with CD that are related to invasive E. coli. TNF is transcribed and translated from scratch following macrophage activation, indicating that macrophages are still active despite many intracellular bacteria. The persistent proliferation of internal bacteria within the phagosomes causes continuous activation and TNF release 42.
Pathogenic Properties of E. Coli
Isolated E. coli bacteria from IBD patients are clonally heterogeneous, belonging to multiple serotypes and sequence types. Though a close genetic hereditary link has been found in pediatric patients with IBD, the concept of IBD that is caused by a specific clone has been generally dismissed 43. In contrast, E. coli is identified in patients with IBD in combination with extraintestinal pathogenic E. Coli; typically B2 and D phylotypes. Extraintestinal pathogenic E. coli. Many studies show that B2 and D phylotypes colonize IBD patients more than healthy people, whereas some other studies suggest that IBD and healthy participants have similar phylogroup distributions 44, 45.These differences in findings could be a product of the different types of samples studied, since it has been observed that transitory E. coli, the most abundant type discovered in stools, B1 and A phylotypes specifically, are commonly found in healthy people. By contrast, resident E. coli, which is the most common identified from biopsy, results mainly D and B2 phylotypes, which are generally associated with IBD. As a result, investigation depending on biopsies samples will tend to show B2 and D strain abundance, even in healthy people. Another factor that alters perceived abundance of phylotypes in IBD is illness severity46. A higher fraction of B2 and D separate has been detected in inactive IBD patients, which has been linked to inflammatory level of tissue 44, 47. This indicates a change in abundance of E. coli toward separates that are more suited to inflamed tissue in IBD patients or are active in the inflammatory process 48. To the present authors’ knowledge, there have been no reports of phylotype distribution discrepancies among UC and CD patient E. coli strains that have distinct sets of virulence genes than pathogenic E. coli (ExPEC) strains. Nevertheless, intestine ExPEC is very rare or perhaps nonexistent49. The presence of virulence factors in E. coli in healthy people is thought to be crucial for the effectiveness of colonization in the gut mucosa. Malignancy gene profiles are inextricably related to the evolutionary origin of the strain47. Where B2 and D are predominant in patients with IBD, more commonly discovered virulence-linked genes indicative of ExPEC were found in IBD patients than in controls, depending on the abundance of phylogenetic groups, and without differences in other investigation types50. A shift in the phylotype distribution would result in increased abundance of E. coli with colonization factors, facilitating fixation and persistence in IBD patients. However, it is unknown if the alteration occurs only in IBD patients or is a universal trend in industrialized nations. Although inspecific genetic characteristics separate E. coli from the UC or CD gut mucosa, certain virulence factors have been discovered that are distributed variably amongst these IBD types. For instance, diarrhea-linked hemolytic E. coli, also known as cell-detaching E. coli (CDEC), has been detected in about 24 % of UC E. coli patients, while detected in only 4.7 % of CD E. coli patients 50. Diarrhea-linked hemolytic E. coli usually carries pilus P, hemolysin, S-fimbria genes, and cytotoxic necrotizing factor 1. The uropathogenic-specific protein (USP) gene, which codes for the uropathogenic-specific protein discovered in UC patient E. coli, is more abundant than in CD patient E. coli 50. The E. coli containing iro gene, which codes for an iron-chelating siderophores receptor, was newly seen to be more very often separated from inflammatory and uninflamed mucosa-inactive patients with UC. Darfeuille-Michaud et al. found a novel pathotype of E. coli with different phenotypic pathogenic characteristics that were not linked with UC but associated with CD, and termed adherent invasive E. coli (AIEC) 51.
Conclusion
IBDs are becoming more common throughout the world, in developing and developed countries. Although the causes are undetermined, a burgeoning body of research has found a clear link between IBD morbidity primarily due to hereditary susceptibility, although in connection with environmental factors, and have shown key characteristics and biomarkers of the gut microbiota that characterize pathogenesis and susceptibility. Variables such as nutrition, tissue damage linked with immune system disturbances, and aberrant gut microbiota have been implicated. E . coli has been found to be an essential microorganism of a healthy gut but can have a key role in the development of IBD.
Acknowledgment
We would like to extend our thanks to everyone that assisted in the English language revision and editing and proofreading of the manuscript.
Conflict of Interests
The authors declare no conflicts of interest.
Funding Source
No funding source
References
- Barnich N, Carvalho FA, Glasser A-L, Darcha C, Jantscheff P, Allez M, et al. CEACAM6 acts as a receptor for adherent-invasive E. coli, supporting ileal mucosa colonization in Crohn disease. The Journal of clinical investigation. 2007;117(6):1566-74.
CrossRef - Erjavec MS. Introductory Chapter: The Versatile Escherichia coli. The Universe of Escherichia coli. 2019:3.
- Baumgart M, Dogan B, Rishniw M, Weitzman G, Bosworth B, Yantiss R, et al. Culture independent analysis of ileal mucosa reveals a selective increase in invasive Escherichia coli of novel phylogeny relative to depletion of Clostridiales in Crohn’s disease involving the ileum. The ISME journal. 2007;1(5):403-18.
CrossRef - Swidsinski A, Ladhoff A, Pernthaler A, Swidsinski S, Loening–Baucke V, Ortner M, et al. Mucosal flora in inflammatory bowel disease. Gastroenterology. 2002;122(1):44-54.
CrossRef - Ng SC, Shi HY, Hamidi N, Underwood FE, Tang W, Benchimol EI, et al. Worldwide incidence and prevalence of inflammatory bowel disease in the 21st century: a systematic review of population-based studies. The Lancet. 2017;390(10114):2769-78.
CrossRef - Loftus E, Harewood G, Loftus C, Tremaine W, Harmsen W, Zinsmeister A, et al. PSC-IBD: a unique form of inflammatory bowel disease associated with primary sclerosing cholangitis. Gut. 2005;54(1):91-6.
CrossRef - M’Koma AE. Inflammatory bowel disease: an expanding global health problem. Clinical medicine insights Gastroenterology. 2013;6:33-47.
CrossRef - Philpott DJ, Sorbara MT, Robertson SJ, Croitoru K, Girardin SE. NOD proteins: regulators of inflammation in health and disease. Nature reviews Immunology. 2014;14(1):9-23.
CrossRef - Cleynen I, Boucher G, Jostins L, Schumm LP, Zeissig S, Ahmad T, et al. Inherited determinants of Crohn’s disease and ulcerative colitis phenotypes: a genetic association study. The Lancet. 2016;387(10014):156-67.
CrossRef - Strober W, Fuss I, Mannon P. The fundamental basis of inflammatory bowel disease. The Journal of clinical investigation. 2007;117(3):514-21.
CrossRef - Fuss IJ, Heller F, Boirivant M, Leon F, Yoshida M, Fichtner-Feigl S, et al. Nonclassical CD1d-restricted NK T cells that produce IL-13 characterize an atypical Th2 response in ulcerative colitis. The Journal of clinical investigation. 2004;113(10):1490-7.
CrossRef - Gilbert JA, Blaser MJ, Caporaso JG, Jansson JK, Lynch SV, Knight R. Current understanding of the human microbiome. Nature medicine. 2018;24(4):392-400.
CrossRef - Coufal S, Galanova N, Bajer L, Gajdarova Z, Schierova D, Jiraskova Zakostelska Z, et al. Inflammatory Bowel Disease Types Differ in Markers of Inflammation, Gut Barrier and in Specific Anti-Bacterial Response. Cells. 2019;8(7).
CrossRef - Zhang M, Sun K, Wu Y, Yang Y, Tso P, Wu Z. Interactions between Intestinal Microbiota and Host Immune Response in Inflammatory Bowel Disease. Front Immunol. 2017;8:942-.
CrossRef - Zhang Y-J, Li S, Gan R-Y, Zhou T, Xu D-P, Li H-B. Impacts of gut bacteria on human health and diseases. International journal of molecular sciences. 2015;16(4):7493-519.
CrossRef - Clapp M, Aurora N, Herrera L, Bhatia M, Wilen E, Wakefield S. Gut microbiota’s effect on mental health: The gut-brain axis. Clin Pract. 2017;7(4):987-.
CrossRef - Khan I, Ullah N, Zha L, Bai Y, Khan A, Zhao T, et al. Alteration of Gut Microbiota in Inflammatory Bowel Disease (IBD): Cause or Consequence? IBD Treatment Targeting the Gut Microbiome. Pathogens (Basel, Switzerland). 2019;8(3):126.
CrossRef - Belkaid Y, Hand TW. Role of the microbiota in immunity and inflammation. Cell. 2014;157(1):121-41.
CrossRef - Duchmann R, Kaiser I, Hermann E, Mayet W, Ewe K, BÜSCHENFELDE KHMZ. Tolerance exists towards resident intestinal flora but is broken in active inflammatory bowel disease (IBD). Clinical & Experimental Immunology. 1995;102(3):448-55.
CrossRef - Zhang M, Sun K, Wu Y, Yang Y, Tso P, Wu Z. Interactions between Intestinal Microbiota and Host Immune Response in Inflammatory Bowel Disease. Front Immunol. 2017;8:942.
CrossRef - Eun CS, Kwak M-J, Han DS, Lee AR, Park DI, Yang S-K, et al. Does the intestinal microbial community of Korean Crohn’s disease patients differ from that of western patients? BMC gastroenterology. 2016;16(1):1-11.
CrossRef - Wang W, Chen L, Zhou R, Wang X, Song L, Huang S, et al. Increased proportions of Bifidobacterium and the Lactobacillus group and loss of butyrate-producing bacteria in inflammatory bowel disease. Journal of clinical microbiology. 2014;52(2):398-406.
CrossRef - Schwiertz A, Jacobi M, Frick J-S, Richter M, Rusch K, Köhler H. Microbiota in pediatric inflammatory bowel disease. The Journal of pediatrics. 2010;157(2):240-4. e1.
CrossRef - Martin HM, Campbell BJ, Hart CA, Mpofu C, Nayar M, Singh R, et al. Enhanced Escherichia coli adherence and invasion in Crohn’s disease and colon cancer. Gastroenterology. 2004;127(1):80-93.
CrossRef - Glassner KL, Abraham BP, Quigley EM. The microbiome and inflammatory bowel disease. Journal of Allergy and Clinical Immunology. 2020;145(1):16-27.
CrossRef - Khan I, Ullah N, Zha L, Bai Y, Khan A, Zhao T, et al. Alteration of gut microbiota in inflammatory bowel disease (IBD): cause or consequence? IBD treatment targeting the gut microbiome. Pathogens. 2019;8(3):126.
CrossRef - Henson MA, Phalak P. Microbiota dysbiosis in inflammatory bowel diseases: in silico investigation of the oxygen hypothesis. BMC systems biology. 2017;11(1):1-15.
CrossRef - Pakbin B, Brück WM, Rossen JW. Virulence Factors of Enteric Pathogenic Escherichia coli: A Review. International journal of molecular sciences. 2021;22(18):9922.
CrossRef - Lopez-Siles M, Martinez-Medina M, Busquets D, Sabat-Mir M, Duncan SH, Flint HJ, et al. Mucosa-associated Faecalibacterium prausnitzii and Escherichia coli co-abundance can distinguish Irritable Bowel Syndrome and Inflammatory Bowel Disease phenotypes. International journal of medical microbiology : IJMM. 2014;304(3-4):464-75.
CrossRef - Sha S, Xu B, Wang X, Zhang Y, Wang H, Kong X, et al. The biodiversity and composition of the dominant fecal microbiota in patients with inflammatory bowel disease. Diagnostic microbiology and infectious disease. 2013;75(3):245-51.
CrossRef - Nataro JP, Kaper JB. Diarrheagenic escherichia coli. Clinical microbiology reviews. 1998;11(1):142-201.
CrossRef - Rehman A, Lepage P, Nolte A, Hellmig S, Schreiber S, Ott SJ. Transcriptional activity of the dominant gut mucosal microbiota in chronic inflammatory bowel disease patients. Journal of medical microbiology. 2010;59(9):1114-22.
CrossRef - Martinez-Medina M, Aldeguer X, Lopez-Siles M, González-Huix F, López-Oliu C, Dahbi G, et al. Molecular diversity of Escherichia coli in the human gut: new ecological evidence supporting the role of adherent-invasive E. coli (AIEC) in Crohn’s disease. Inflammatory bowel diseases. 2009;15(6):872-82.
CrossRef - Mylonaki M, Rayment NB, Rampton DS, Hudspith BN, Brostoff J. Molecular characterization of rectal mucosa-associated bacterial flora in inflammatory bowel disease. Inflamm Bowel Dis. 2005;11(5):481-7.
CrossRef - Kotlowski R, Bernstein CN, Sepehri S, Krause DO. High prevalence of Escherichia coli belonging to the B2+D phylogenetic group in inflammatory bowel disease. Gut. 2007;56(5):669-75.
CrossRef - Gullberg E, Söderholm JD. Peyer’s patches and M cells as potential sites of the inflammatory onset in Crohn’s disease. Annals of the New York Academy of Sciences. 2006;1072(1):218-32.
CrossRef - Martin HM, Campbell BJ, Hart CA, Mpofu C, Nayar M, Singh R, et al. Enhanced Escherichia coli adherence and invasion in Crohn’s disease and colon cancer. Gastroenterology. 2004;127(1):80-93.
CrossRef - Boudeau J, Glasser A-L, Masseret E, Joly B, Darfeuille-Michaud A. Invasive ability of an Escherichia coli strain isolated from the ileal mucosa of a patient with Crohn’s disease. Infection and immunity. 1999;67(9):4499-509.
CrossRef - Mimouna S, Gonçalvès D, Barnich N, Darfeuille-Michaud A, Hofman P, Vouret-Craviari V. Crohn disease-associated Escherichia coli promote gastrointestinal inflammatory disorders by activation of HIF-dependent responses. Gut microbes. 2011;2(6):335-46.
CrossRef - Bringer M-A, Rolhion N, Glasser A-L, Darfeuille-Michaud A. The oxidoreductase DsbA plays a key role in the ability of the Crohn’s disease-associated adherent-invasive Escherichia coli strain LF82 to resist macrophage killing. Journal of bacteriology. 2007;189(13):4860-71.
CrossRef - Navarre WW, Zychlinsky A. Pathogen‐induced apoptosis of macrophages: a common end for different pathogenic strategies: Microreview. Cellular microbiology. 2000;2(4):265-73.
CrossRef - Ryan P, Kelly RG, Lee G, Collins JK, O’sullivan GC, O’connell J, et al. Bacterial DNA within granulomas of patients with Crohn’s disease—detection by laser capture microdissection and PCR. Official journal of the American College of Gastroenterology| ACG. 2004;99(8):1539-43.
CrossRef - Sepehri S, Khafipour E, Bernstein CN, Coombes BK, Pilar AV, Karmali M, et al. Characterization of Escherichia coli isolated from gut biopsies of newly diagnosed patients with inflammatory bowel disease. Inflammatory bowel diseases. 2011;17(7):1451-63.
CrossRef - Dreux N, Denizot J, Martinez-Medina M, Mellmann A, Billig M, Kisiela D, et al. Point mutations in FimH adhesin of Crohn’s disease-associated adherent-invasive Escherichia coli enhance intestinal inflammatory response. PLoS pathogens. 2013;9(1):e1003141.
CrossRef - Fang X, Monk JM, Mih N, Du B, Sastry AV, Kavvas E, et al. Escherichia coli B2 strains prevalent in inflammatory bowel disease patients have distinct metabolic capabilities that enable colonization of intestinal mucosa. BMC systems biology. 2018;12(1):1-10.
CrossRef - Raisch J, Buc E, Bonnet M, Sauvanet P, Vazeille E, de Vallée A, et al. Colon cancer-associated B2 Escherichia coli colonize gut mucosa and promote cell proliferation. World Journal of Gastroenterology: WJG. 2014;20(21):6560.
CrossRef - Kouadio-Ngbesso N, Atobla K, Attien PY, Kouame-Sina M, Koffi RA, Adingra AA, et al. Comparative Biotypic and Phylogenetic Profiles of <i>Escherichia coli</i> Isolated from Resident Stool and Lagoon in Fresco (Côte d’Ivoire). International Journal of Microbiology. 2019;2019:9708494.
CrossRef - Elliott TR, Hudspith BN, Wu G, Cooley M, Parkes G, Quiñones B, et al. Quantification and characterization of mucosa-associated and intracellular Escherichia coli in inflammatory bowel disease. Inflamm Bowel Dis. 2013;19(11):2326-38.
CrossRef - Vejborg RM, Hancock V, Petersen AM, Krogfelt KA, Klemm P. Comparative genomics of Escherichia coli isolated from patients with inflammatory bowel disease. BMC genomics. 2011;12:316.
CrossRef - Čurová K, Kmeťová M, Sabol M, Gombošová L, Lazúrová I, Siegfried L. Enterovirulent E. coli in inflammatory and noninflammatory bowel diseases. Folia microbiologica. 2009;54(1):81-6.
CrossRef - Pilarczyk-Zurek M, Chmielarczyk A, Gosiewski T, Tomusiak A, Adamski P, Zwolinska-Wcislo M, et al. Possible role of Escherichia coli in propagation and perpetuation of chronic inflammation in ulcerative colitis. BMC gastroenterology. 2013;13(1):1-11.
CrossRef

This work is licensed under a Creative Commons Attribution 4.0 International License.





