Manuscript accepted on : 21-08-2020
Published online on: 10-09-2020
Plagiarism Check: Yes
Reviewed by: Shulbhi V
Second Review by: Kandiah Pakeerathan ![]()
1Department of Biological Sciences, Faculty of Science, King Abdulaziz University (KAU), P.O. Box 80141, Jeddah 21589, Saudi Arabia
Corresponding Author E-mail : rmakki@kau.edu.sa
DOI : http://dx.doi.org/10.13005/bbra/2858
ABSTRACT: Salt stress is among environmental conditions that severely retards plant growth. Scope of this work is the detection of transcription factors that might participate in regulating salt-stressed genes in wild barley (Hordeum spontaneum). Expression profiles of important types of transcription factors (TFs) were displayed. They include WRKY and MYB, that were regulated under salt stress. WRKY19 and NAC96 are known to induce stress tolerance through activation of DREB2A (or Ap2-ERF). NAC96 concordantly upregulated with DREB2A gene under salt stress in H. spontaneum, a possible crosstalking to compensate the negative performance of WRKY19 gene. P5CS, for proline accumulation, is also known to be driven by ERF1 and genes encoding these proteins concordantly upregulated in H. spontaneum under salt stress supporting NAC96/ERF1/P5CS crosstalking towards proline accumulation under stress. Genes encoding enzymes participating in the last steps of glucose, sucrose and maltose biosyntheses concordantly upregulated with WRKY11 that is also involved in driving genes encoding free proline. B-box zinc finger protein 21 (BZF21) concordantly expressed with genes encoding catalase and SAUR40 indicating that BZF21 gene might drive expression of the two genes under salt stress. Upregulated WRKY41 and WRKY46 under salt stress in wild barley are known to exhibit enhanced stomatal closure, reactive oxygen species (ROS) scavenging, lateral roots development via regulation of ABA signaling and auxin homeostasis. The latter action is governed by GH3.8 gene that was upregulated in wild barley. MYB30 is known for being SUMOylated by SIZ1. In the present study, MYB30, MYB44 and MYB3R-2 genes were concordantly expressed with SIZ2 gene supporting their crosstalking under salt stress in H. spontaneum. Based on the regulation of WRKY19 and MYB30 genes under salt stress in H. spontaneum, we suggest that the first is a positive activator, while the second is a negative activator of FT gene that drives early flowing in plants. MYB44 that promotes stomatal closure under stress can also serve in conferring tolerance to abiotic stresses in wild barley. Several other downregulated genes under salt stress, e.g., MYB1, MYB20 and MYB73, were previously reported to negatively regulate abiotic stress tolerance in plants. We suggest that WRKY gene family participates in salt stress responses in leaves of H. spontaneum following approaches different from those of other plants. Regulation of MYB gene family is almost similar to that of other plant species under salt stress. In conclusion, the present study addresses some of the regulatory frameworks driving expression of salt-related genes in H. spontaneum that can be utilized in plant, e,g, cereals, breeding programs to improve their salt stress tolerance.
KEYWORDS: DREB; FT; GH3; MYB; NAC; SAUR, SIZ; WRKY
Download this article as:| Copy the following to cite this article: Makki. R. M. Molecular Networking of Regulated Transcription Factors under Salt Stress in Wild Barley (H. spontaneum). Biosci Biotech Res Asia 2020;17(3). |
| Copy the following to cite this URL: Makki. R. M. Molecular Networking of Regulated Transcription Factors under Salt Stress in Wild Barley (H. spontaneum). Biosci Biotech Res Asia 2020;17(3). Available from: https://bit.ly/32jPcnT |
Introduction
Salt stress is one of the most devastating environmental conditions that extremely restrict plant growth and yield. For a plant to survive such harsh condition, a series of tolerance mechanisms can occur to help plant adapt and respond properly to this condition.1 Earlier reports indicate that expression levels of different stress-related genes are regulated by transcription factors (TFs) that work as stimulators of individual genes or act as master switches driving a battery of genes or a whole pathway such as stress signal transduction pathways.2-8 WRKY is among the largest families of TFs that play important roles in modulating physiological processes in plants under stress conditions.8-12 Protein encoded by this TF gene family is characterized by a 60 amino acids domain of highly conserved WRKYGQK heptapeptide at the N-terminal and an atypical zinc finger-like motif at its C-terminal.13, 14 This family contains over 70 members in Arabidopsis.13, 15 55 in cucumber,16 119 in maize,17 94 in barley,18 and 100 in rice.19 Encoded proteins of this family are characterized by a 60-amino acids domain containing the WRKY amino acid sequence at its amino-terminal end and a putative zinc finger motif at its carboxy-terminal end. Based on number and diversity of WRKY domains, WRKY proteins are classified into three groups (I, II and III) of which category I proteins harbor two domains, while proteins of groups II and III harbor only one domain. Groups II and III proteins differ in zinc finger structures (C2H2 in group II, while C2HC in group III) (14, 20). Previous study reported a number of 74 WRKY proteins in Arabidopsis, while over 100 in rice (Oryza sativa) (10). WRKY TFs have specificity to bind W-box [TTGAC(C/T)] of promoters of their target genes, which subsequently wire genetic circuits towards downstream biological responses.20, 21
WRKY TFs can either negatively or positively trigger a certain response under stress conditions.19 These regulation patterns as well as members participating in a given condition can changes from a plant to the other. For example, WRKY54 and WRKY70 in Arabidopsis negatively regulate leaf senescence.22 While, WRKY23 positively enhanced pathogen defense and overexpression of maize WRKY58 in rice.23 and wheat WRKY1 and WRKY33 in Arabidopsis positively conferred drought and salt tolerance.24 WRKY TFs also reported to be involved in abiotic stress by wiring ABA signaling pathway.25 For example, Chrysanthemum WRKY1 enhanced abiotic stress tolerance, while cotton WRKY17 overexpressed in tobacco reduced tolerance by regulating a number of genes in ABA signaling pathway and reactive oxygen species (ROS) production.
MYB is also a family of TFs involved in response to abiotic stresses in plants.26 Of which, expression of MYB108 gene in Arabidopsis is induced in response to salt stress and participate in crosstalking between abiotic and biotic stresses via orchestration of signaling pathways of jasmonic acid (JA) and gibberellic acid (GA) 27, 28 While, MYB65 participates in GA signaling in growth and flowering processes.29 Our work also showed that the expression of this gene increased in roots in response to both stresses whereas, in leaves up-regulated only in response to salt stress. Expression of genes encoding MYB differs in different tissues in response to salt stress as MYB34 in Arabidopsis, for example, is normally upregulated in root tissue and its expression in leaves increases only in response to stress, whereas MYB47 and MYB32 were common in both tissues.28, 30
In the present study, we have demonstrated important types of TFs including WRKY and MYB that were regulated under salt stress in wild barley Hordeum spontaneum. The information recovered from this work can be helpful in improving plant salt stress tolerance in the future.
Materials and Methods
Salt stress experiment was conducted on H. spontaneum as previously described.31 Fourteen-day-old seedlings were treated with salt (500 mM NaCl) and total RNAs were harvested in a replicated experiment from leaves at 0 (control), 2, 12 and 24 h time point using Trizol (Invitrogen, Life Tech, Grand Island, NY, USA). Then, RNAs were treated with RNase-free DNase (Promega Corporation, Madison, WI, USA) and 1 U/ul of RNasin® Plus RNase Inhibitor as described (Promega Corporation, Madison, WI, USA). Total RNA samples were, then, shipped to Beijing Genomics Institute (BGI), Shenzhen, China for deep sequencing using illumina Miseq. Generated raw data were retrieved in FASTQ format and submitted to the NCBI and experiment received accession number of PRJNA227211 (https://www.ncbi.nlm.nih.gov/ bioproject?LinkName=sra_bioproject&from_uid=537429). Individual accession numbers of raw data of different samples are available in NCBI (https://www.ncbi.nlm.nih.gov/ sra?LinkName=bioproject_sra_all&from_uid=227211). Raw data was processed as described32 and clean data was subjected to genome-guided Trinity de novo transcriptome assembly (https://github.com/trinityrnaseq/trinityrnaseq/wiki/Genome-Guided-Trinity-Transcriptome-Assembly) with Hordeum vulgare genome (https://plants.ensembl.org/ Hordeum_vulgare/Info/Index, Taxonomy ID 112509) used as the guide. Differential expression and cluster analysis were done by EdgeR (version 3.0.0, R version 2.1.5) with proper algorism and fold change values of ≥ 2 measured against actin house-keeping gene. Annotation of the recovered transcripts was done using Blast2GO (http://www.blast2go.org/). Subsequent bioinformatics approach was done as described.33 Predicted CDSs were annotated against protein database in order to assign functions of transcripts. Protein domains common in TFs were identified using HMMER3 software.34
Then, RNA-Seq datasets were validated via qRT-PCR of four randomly selected genes using the Agilent Mx3000P qPCR Systems (Agilent technology, USA) as previously described.31 Transcripts selected from cluster analysis were upregulated at 2 and 12 h time points. Primer sequences are shown in Table S1. Calculations referring to expression levels of each transcript were done relative to that under control condition and barley actin gene was used as the house-keeping gene.
Results and Discussion
For validating RNA-Seq datasets, qRT-PCR was done for four randomly selected transcripts encoding transcripts that were either upregulated at 2 and 12 h time points, upregulated at 2 h time point, or downregulated at 2 and 12 h time points of salt stress and results aligned with RNA-Seq datasets for transcripts used for validation (Figure S1). Cluster analysis resulted in the recovery of over 10000 differentially expressed (DE) transcripts with fold change of ≥ 2 under salt stress including over 600 TFs highlighted in Table S2 that are separately shown in Table S3. Well-known TF families for their response to abiotic stresses include WRKY and MYB. Genes encoding WRKY activated at 2 and 12 h time points under salt stress include WRKY2, WRKY11, WRKY41, WRKY46, WRKY50 and WRKY71 (Figure 1a). Gene encoding WRKY24 was upregulated at 2 h time point only, while downregulated at 24 h time point (Figure 1c). Downregulated WRKY genes in the present study include WRKY19 and WRKY35 (Figure 1b). Genes encoding MYB under salt stress include MYB30, MYB44, MYB62, MYB3R-2 and MYB3R-4, while downregulated MYB genes include MYB1, MYB20, MYB73 and MYBS3 (Figure 2).
WRKY2 and WRKY19 were reported by Niu et al.35 to induce stress tolerance in wheat through activation of STZ (salt tolerance zinc finger) and DREB2A (dehydration-responsive element binding 2A) pathways, respectively. Although a large number of zinc finger genes11 in the present study was regulated in leaves H. spontaneum under salt stress (Table S3), STZ gene was not regulated. Then, STZ gene cannot be used in tracing regulation of WRKY2 gene. WRKY19 gene was downregulated in leaves of H. spontaneum under salt stress (Figure 1b), thus, no activation of DREB2A gene is expected. DREB2A is among AP2-ERF (Apetala2/Ethylene responsive factor) gene family and a recent report indicated that DREB2A is also affected by other TFs, ex., NAC96.28 Interestingly, two Ap2-ERF gene isoforms and a gene encoding NAC96 were upregulated in cluster 1 under salt stress in leaves of H. spontaneum (Figure 3 and Table S2) indicating that upregulation of Ap2-ERF gene can compensate the negative regulation of WRKY19 gene in H. spontaneum.
 |
Figure 1: Up- (a), downregulated (b) and up-/ downregulated (c) transcripts of WRKY family under salt stress (1 M NaCl) across 0, 2, 12 and 24 h time points in leaves of H. |
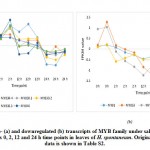 |
Figure 2: Up- (a) and downregulated (b) transcripts of MYB family under salt stress (1 M NaCl) across 0, 2, 12 and 24 h time points in leaves of H. spontaneum. Original RNA-Seq data is shown in Table S2. |
 |
Figure 3: Expression pattern of transcripts encoding two DREB2A (AP2-ERF) isoforms under salt stress (1 M NaCl) across 0, 2, 12 and 24 h time points in leaves of H. factor. |
Overexpression of WRKY2 gene in grapevine increased proline under salt stress.36 Two other recent reports indicated that WRKY2 in wheat37 and WRKY11 in soybean38 also drive genes encoding free proline and soluble sugars under drought stress. Proline and sugars are known as important osmolytes that neutralize the effects of salt and alleviate stress.39, 40 Interestingly, gene encoding Delta-1-pyrroline-5-carboxylate synthase (P5CS) for proline accumulation was concordantly upregulated under salt stress with that encoding ERF1 (ethylene responsive factor 1) gene, other DREB gene derivative, in cluster 32 (Figure 4 and Table S2). Then, proline accumulation due to the function of P5CS gene in H. spontaneum under salt stress can also be driven by ERF1 gene that is likely controlled by NAC96, not by either WRKY2 or WRKY11. As per expected sugar levels under salt stress in H. spontaneum, results indicated that genes encoding enzymes participating in the last step of glucose (e.g., beta-glucosidase), sucrose (e.g., sucrose synthase 6) and maltose (e.g., beta-amylase 8) biosynthesis concordantly upregulated with WRKY2 in clusters 1 and 5, while WRKY11 and MYB3R-2 in cluster 6 (Table S2) under salt stress (Figure 5). Accordingly, we speculate that WRKY 2 and WRKY11 are involved in driving genes encoding soluble sugars as two different mechanisms of salt stress tolerance in H. spontaneum.
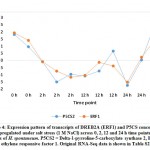 |
Figure 4: Expression pattern of transcripts of DREB2A (ERF1) and P5CS concordantly upregulated under salt stress (1 M NaCl) across 0, 2, 12 and 24 h time pointsClick here to View Figure |
 |
Figure 5: Expression pattern of transcripts encoding SS6, BA8, BG25, WRKY2, WRKY11 and MYB3R-2 concordantly upregulated under salt stress (1 M NaCl) across 0, 2, 12 and 24 h time points in leaves of H. spontaneum.Click here to View Figure |
Recently, WRKY11 was also proven to induce elevated levels of superoxide dismutase (SOD) and catalase in soybean.38 In the present study, upregulated WRKY11 gene does not seem to concordantly express with SOD regulated gene isoforms in H. spontaneum, where SOD gene isoforms were downregulated (Figure 6 and Table S2) as shown in clusters 2 and 27 and no other TF can likely complement WRKY11 effect whose upregulation pattern of its three isoforms in H. spontaneum was different (cluster 1). Although both genes are upregulated, WRKY11 gene does not either concordantly express with isoforms of gene encoding catalase (existing in cluster 23), but gene encoding another TF namely B-box zinc finger protein 21 (BZF21) concordantly expressed with the two isoforms of gene encoding catalase, thus, possibly drive expression of this gene in H. spontaneum instead of WRKY11 (Figure 7 and Table S2). B-box zinc finger proteins were reported to enhance salt and drought stresses tolerance in Arabidopsis (Liu et al., 2019). Interestingly, gene encoding SAUR40 also concordantly expressed with BZF21 and catalase genes in cluster 23 (Figure 7 and Table S2). SAUR40 gene is among a family acting as a regulator of cell elongation and plant growth performance41 and a stimulator of shoot elongation due to auxin signaling.42 Thus, we speculate that BZF21 might drive expression of both catalase and SAUR40 genes as genes encoding the three metabolites are concordantly expressed (Figure 7).
Participation of the two TFs, namely WRKY24 and WRKY71, as responsive elements under salt stress was argued in rice.43 However, expression patterns of isoforms of these two TFs seem to be controversial (Figure 1) as gene encoding the first was upregulated at 2 h time point and downregulated at 24 h time point, while gene encoding the second was upregulated at 2 and 12 time points. Xie et al. (19) indicated that WRKY24 gene is induced by ABA signaling, while Basu and Roychoudhury43 indicated that ABA signaling induces higher expression of WRKY71 gene and many other TFs. Interestingly, the authors indicated that WRKY24 gene showed expression even lower than that of the control untreated samples under salt stress. WRKY71 was recently reported to antagonistically act against both salt-delayed flowering and escaping salt stress in Arabidopsis through the induction of gene encoding FLOWERING LOCUS T (FT) (44). Surprisingly, two isoforms of the latter gene in clusters 4 and 8 seem concordantly downregulated with gene encoding WRKY19 of cluster 4 rather than with gene encoding WRKY71 of cluster 21 (Figure 8 and Table S2). We cannot jump to conclusions on the relationship between WRKY19 and FT genes unless an experiment to detect the consequences of WRKY19 gene being knocked out in Arabidopsis model.
 |
Figure 6: Expression pattern of transcripts encoding SOD downregulated under salt stress (1 M NaCl) across 0, 2, 12 and 24 h time points in leaves of H. spontaneum |
 |
Figure 7: Expression pattern of transcripts encoding isoforms of catalase concordantly upregulated with BZF21 and SAUR genes |
Aligning with the results of the present study, WRKY41 and WRKY46 were reported to positively relate to salt stress tolerance in tobacco.45 Genes encoding the two TFs were upregulated under salt stress in H. spontaneum as shown in clusters 11 and 1, respectively (Figure 1 and Table S2). Overexpressing the cotton WRKY41 gene in tobacco exhibited enhanced stomatal closure and reactive oxygen species (ROS) scavenging when plants were exposed to osmotic stress.45 WRKY46 acts in Arabidopsis in developing lateral roots under osmotic/salt stress via regulation of ABA signaling and auxin homeostasis.46 Auxin homeostasis is known to be regulated by GRETCHEN HAGEN3 or GH3 gene family in “Plant hormone signal transduction” pathway.42 In the present study, GH3.8 gene was upregulated in cluster 17 with no exact TF concordantly expressed with it (Figure 9 and Table S2). No conclusive information on the function of WRKY50 (cluster 1) is available except that it acts as a positive regulator in the salicylic acid (SA) signaling pathway and probably ABA signaling pathway in Arabidopsis, while a negative regulator in jasmonic acid (JA) signaling.47, 48 Very little is known about the mode of action of WRKY35 except that its expression participates in conferring salt stress tolerance in zoysia grass.49 We conclude that WRKY gene family participates in salt stress responses in leaves of H. spontaneum in ways different from those in other plant species.
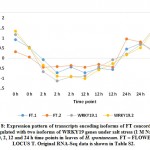 |
Figure 8: Expression pattern of transcripts encoding isoforms of FT concordantly upregulated with two isoforms of WRKY19 genes under salt stress |
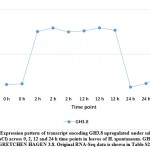 |
Figure 9: Expression pattern of transcript encoding GH3.8 upregulated under salt stress (1 M NaCl) across 0, 2, 12 and 24 h time points in leaves of H. spontaneum. |
Expression of five MYB genes, namely MYB30, MYB44, MYB62, MYB3R-2 and MYB3R-4, was proven to be increased under salt stress in leaves of H. spontaneum (Figure 2a), while expression of four, namely MYB1, MYB20, MYB73 and MYBS3, was decreased (Figure 2b). MYB30, an R2R3‐MYB TF, was studied by Gong et al.50 and results indicated that expression in the perennial wall-rocket (Diplotaxis tenuifolia L.) increased under salt stress up to 4 h time point, while gradually decreased up to 24 h time point in perfect alignment with results of the present study with regard to regulation of gene encoding this TF. MYB30 was proven to be SUMOylated by SIZ150, 51 SUMOylation represents a post-translational regulation involved in various cellular processes including response to stresses.52 Small Ubiquitin-like Modifier (SUMO) proteins, like SIZ1 and SIZ2, represent a family of small proteins covalently attached to a certain protein, while detached from others to modify target protein’s (e.g., MYB30) function. In the present study, two isoforms of MYB30 gene as well as MYB44 and MYB3R-2 genes concordantly expressed with SIZ2 gene in cluster 6 under salt stress in H. spontaneum (Figure 10 and Table S2). MYB30 also accelerates flowering both in long and short days. Early flowering is mediated by elevated expression of FLOWERING LOCUS T (FT) gene that is mainly activated by CONSTANS (CO). However, MYB30 can also drive expression of FT gene,53 a phenomenon that we speculated for WRKY19 gene under salt stress in H. spontaneum. The major difference between the possible regulation of WRKY19 or MYB30 gene is that the first is a positive activator, while the second is a negative activator of FT gene. This controversial speculated regulation of WRKY19 and MYB30 genes under salt stress in H. spontaneum is shown in Figure 11. MYB30 was also reported to participate in ABA signaling response (51), in accumulation of very-long-chain fatty acids such as waxes, phospholipids, and complex sphingolipids,54 and in promoting the expression of a subset of brassinosteroids (BRs) target genes.55, 56 No results were detected on the regulation of genes encoding any of the above-mentioned compounds under salt stress in H. spontaneum.
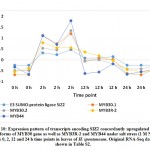 |
Figure 10: Expression pattern of transcripts encoding SIZ2 concordantly upregulated with two isoforms of MYB30 gene as well as MYB3R-2 and MYB44 under salt stress |
Interestingly, MYB44 was proven to be a negative regulator of ABA signaling and abiotic stresses in Arabidopsis,57 while positively increased sensitivity of seed germination to ABA58 The latter authors indicated that phosphorylation of MYB44 by MAPK is mandatory for its function. Nonetheless, Jung et al.59 indicated that MYB44 promotes stomatal closure, a characteristic shared with WRKY41 that consequently serves in conferring tolerance to abiotic stresses in Arabidopsis. Tolerance is conferred for plants overexpressing MYB44 gene because they exhibit a reduced rate of water loss, reduced rate of genes encoding serine/threonine protein phosphatases 2C (PP2Cs), then enhanced tolerance to drought and salt stress. Nonetheless, genes encoding MYB44, protein phosphatase 2C and two isoforms of protein phosphatase 1 (PP1) in the present study are concordantly expressed in cluster 6 (Figure 12 and Table S2). Explanation of concordant expression of MYB44 and genes encoding the two phosphatases might be that MYB44 was upregulated only at 2 h time point only, while the other genes were upregulated also at 12 h time point. This indicates that negative regulation of MYB44 might take place only at 12 h time point.
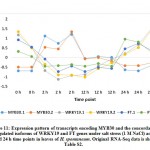 |
Figure 11: Expression pattern of transcripts encoding MYB30 and the concordantly downregulated isoforms of WRKY19 and FT genes under salt stress. |
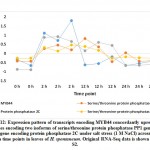 |
Figure 12: Expression pattern of transcripts encoding MYB44 concordantly upregulated with genes encoding two isoforms of serine/threonine protein phosphatase |
(Pi) starvation-induced genes and suppression of gibberellic acid (GA) biosynthesis under nutrient stress. Authors claimed that cross-talking between Pi homeostasis and GA is an adaptive mechanism under abiotic stresses. Therefore, it is logic that MYB62 negatively regulate expression of gibberellin-regulated protein under salt stress in H. spontaneum as previously described.60 In the present study, MYB62 exists in cluster 1 whose expression of transcripts indicated upregulation at 2 and 12 h time points, while one gibberellin-regulated protein exists in cluster 4 whose expression of transcripts indicated downregulation at 2 and 12 h time points (Figure 13 and Table S2). As per MYB3R-4, Haga et al.61 indicated its participation in pleiotropic development and regulation of multiple G2/M-specific genes in Arabidopsis. None of the genes involved in the latter processes were regulated in H. spontaneum under salt stress.
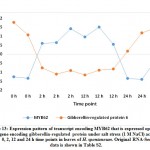 |
Figure 13: Expression pattern of transcript encoding MYB62 that is expressed oppositely to gene encoding gibberellin-regulated protein under salt stress |
MYB1, MYB20, MYB73 and MYBS3 genes were shown to be downregulated under salt stress in H. spontaneum (Figure 2b). These TFs were previously reported to negatively regulate abiotic stress tolerance in plants except for MYBS3 that was reported for its positive role in abiotic stresses, particularly cold stress tolerance in rice via mediation of a-amylase gene expression. 62 In H. spontaneum, MYBS3 seems concordantly expressed with a-amylase gene although the first exists in cluster 4, while the second exists in cluster 2 (Figure 14 and Table S2). Wang et al.63 claimed that MYB1 negatively regulates seed germination under saline conditions in Arabidopsis by regulating the levels of the stress hormone abscisic acid (ABA). Similar conclusions were reached by Gao et al.47 in their work on MYB20 in Arabidopsis with regard to the negative regulation of ABA under drought stress. Loss-of-function experiment of MYB73 gene resulted in drought or/and salt tolerance due to its negative regulation of SOS1 (salt overly sensitive 1) and SOS3 genes.64, 65 No SOS genes are regulated under salt stress in H. spontaneum.
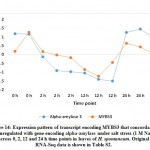 |
Figure 14: Expression pattern of transcript encoding MYBS3 that concordantly downregulated with gene encoding alpha-amylase under salt stress |
 |
Figure 15: Networking of transcription factor regulated in H. spontaneum under salt stress and their known contribution to salt stress tolerance in plants. |
Summary of the overall molecular networking involving transcription factors and their concordantly expressed genes along with downstream biological processes towards conferring salt stress tolerance in H. spontaneum under salt stress is shown in Figure 15.
Conclusion
In conclusion, we suggest that WRKY gene family participates in salt stress responses in leaves of H. spontaneum of which some of them follow different approaches, in terms of their regulation under salt stress as well as the downstream responsive genes, from those of other plant species. Regulation of MYB gene family in H. spontaneum seems similar, to a large extent, to that of other plant species under salt stress. The present study addressed some of the molecular mechanisms by which H. spontaneum follows under salt stress in order to stand severe salt stress. One of the important avenue towards improving salt stress tolerance is understanding the regulatory elements, e.g. transcription factors, that drive important salt-related genes. This information might be useful in subsequent breeding programs in cultivated barley and other cereal crops.
Acknowledgments
The author thanks Professor Dr. Ahmed Bahieldin for providing aliquots of total RNA collected from wild barley at different time points of salt stress for validating RNA-Seq dataset.
Funding Source
There is no funding source.
Conflict of interest
The author declares no conflict of interest.
References
- Ingram J, Bartels D. The molecular basis of dehydration tolerance in plants. Annual review of plant biology. 1996;47(1):377-403.
- Zhu J-K. Salt and drought stress signal transduction in plants. Annual review of plant biology. 2002;53(1):247-73.
- Shinozaki K, Dennis ES. Cell signalling and gene regulation: global analyses of signal transduction and gene expression profiles. Current Opinion in Plant Biology. 2003;6(5):405-9.
- Chen WJ, Zhu T. Networks of transcription factors with roles in environmental stress response. Trends in plant science. 2004;9(12):591-6.
- Yamaguchi-Shinozaki K, Shinozaki K. Organization of cis-acting regulatory elements in osmotic-and cold-stress-responsive promoters. Trends in plant science. 2005;10(2):88-94.
- Xiong L, Schumaker KS, Zhu J-K. Cell signaling during cold, drought, and salt stress. The plant cell. 2002;14(suppl 1):S165-S83.
- Tuteja N. Abscisic acid and abiotic stress signaling. Plant signaling & behavior. 2007;2(3):135-8.
- Budak H, Kantar M, Yucebilgili Kurtoglu K. Drought tolerance in modern and wild wheat. The Scientific World Journal. 2013;2013:548246.
- Ryu H-S, Han M, Lee S-K, Cho J-I, Ryoo N, Heu S, et al. A comprehensive expression analysis of the WRKY gene superfamily in rice plants during defense response. Plant cell reports. 2006;25(8):836-47.
- Eulgem T, Somssich IE. Networks of WRKY transcription factors in defense signaling. Current opinion in plant biology. 2007;10(4):366-71.
- Pandey SP, Somssich IE. The role of WRKY transcription factors in plant immunity. Plant physiology. 2009;150(4):1648-55.
- Li H, Gao Y, Xu H, Dai Y, Deng D, Chen J. ZmWRKY33, a WRKY maize transcription factor conferring enhanced salt stress tolerances in Arabidopsis. Plant Growth Regulation. 2013;70(3):207-16.
- Eulgem T, Rushton PJ, Robatzek S, Somssich IE. The WRKY superfamily of plant transcription factors. Trends in plant science. 2000;5(5):199-206.
- Ülker B, Somssich IE. WRKY transcription factors: from DNA binding towards biological function. Current opinion in plant biology. 2004;7(5):491-8.
- Dong J, Chen C, Chen Z. Expression profiles of the Arabidopsis WRKY gene superfamily during plant defense response. Plant molecular biology. 2003;51(1):21-37.
- Wei K-F, Chen J, Chen Y-F, Wu L-J, Xie D-X. Molecular phylogenetic and expression analysis of the complete WRKY transcription factor family in maize. DNA research. 2012;19(2):153-64.
- Ling J, Jiang W, Zhang Y, Yu H, Mao Z, Gu X, et al. Genome-wide analysis of WRKY gene family in Cucumis sativus. BMC genomics. 2011;12(1):471.
- Liu D, Leib K, Zhao P, Kogel K-H, Langen G. Phylogenetic analysis of barley WRKY proteins and characterization of HvWRKY1 and-2 as repressors of the pathogen-inducible gene HvGER4c. Molecular genetics and genomics. 2014;289(6):1331-45.
- Xie Z, Zhang Z-L, Zou X, Huang J, Ruas P, Thompson D, et al. Annotations and functional analyses of the rice WRKY gene superfamily reveal positive and negative regulators of abscisic acid signaling in aleurone cells. Plant physiology. 2005;137(1):176-89.
- Rushton PJ, Somssich IE, Ringler P, Shen QJ. WRKY transcription factors. Trends in plant science. 2010;15(5):247-58.
- Ciolkowski I, Wanke D, Birkenbihl RP, Somssich IE. Studies on DNA-binding selectivity of WRKY transcription factors lend structural clues into WRKY-domain function. Plant molecular biology. 2008;68(1-2):81-92.
- Besseau S, Li J, Palva ET. WRKY54 and WRKY70 co-operate as negative regulators of leaf senescence in Arabidopsis thaliana. Journal of experimental botany. 2012;63(7):2667-79.
- Cai R, Zhao Y, Wang Y, Lin Y, Peng X, Li Q, et al. Overexpression of a maize WRKY58 gene enhances drought and salt tolerance in transgenic rice. Plant Cell, Tissue and Organ Culture (PCTOC). 2014;119(3):565-77.
- He G-H, Xu J-Y, Wang Y-X, Liu J-M, Li P-S, Chen M, et al. Drought-responsive WRKY transcription factor genes TaWRKY1 and TaWRKY33 from wheat confer drought and/or heat resistance in Arabidopsis. BMC plant biology. 2016;16(1):116.
- Osakabe Y, Osakabe K, Shinozaki K, Tran L-SP. Response of plants to water stress. Frontiers in plant science. 2014;5:86.
- Rasheed S, Bashir K, Matsui A, Tanaka M, Seki M. Transcriptomic analysis of soil-grown Arabidopsis thaliana roots and shoots in response to a drought stress. Frontiers in plant science. 2016;7:180.
- Mengiste T, Chen X, Salmeron J, Dietrich R. The BOTRYTIS SUSCEPTIBLE1 gene encodes an R2R3MYB transcription factor protein that is required for biotic and abiotic stress responses in Arabidopsis. The Plant Cell. 2003;15(11):2551-65.
- Ghorbani R, Alemzadeh A, Razi H. Microarray analysis of transcriptional responses to salt and drought stress in Arabidopsis thaliana. Heliyon. 2019;5(11):e02614.
- Gocal GFW, Sheldon CC, Gubler F, Moritz T, Bagnall DJ, MacMillan CP, et al. GAMYB-like genes, flowering, and gibberellin signaling in Arabidopsis. Plant physiology. 2001;127(4):1682-93.
- Bhargava A, Clabaugh I, To JP, Maxwell BB, Chiang Y-H, Schaller GE, et al. Identification of cytokinin-responsive genes using microarray meta-analysis and RNA-Seq in Arabidopsis. Plant Physiology. 2013;162(1):272-94.
- Bahieldin A, Atef A, Shokry AM, Al-Karim S, Al Attas SG, Gadallah NO, et al. Structural identification of putative USPs in Catharanthus roseus. Comptes rendus biologies. 2015;338(10):643-9.
- Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114-20.
- Bahieldin A, Atef A, Edris S, Gadalla NO, Ramadan AM, Hassan SM, et al. Multifunctional activities of ERF109 as affected by salt stress in Arabidopsis. Scientific reports. 2018;8(1):1-10.
- Eddy SR. A new generation of homology search tools based on probabilistic inference. Genome Informatics 2009: Genome Informatics Series Vol 23: World Scientific; 2009. p. 205-11.
- Niu CF, Wei WEI, Zhou QY, Tian AG, Hao YJ, Zhang WK, et al. Wheat WRKY genes TaWRKY2 and TaWRKY19 regulate abiotic stress tolerance in transgenic Arabidopsis plants. Plant, cell & environment. 2012;35(6):1156-70.
- Mzid R, Zorrig W, Ayed RB, Hamed KB, Ayadi M, Damak Y, et al. The grapevine VvWRKY2 gene enhances salt and osmotic stress tolerance in transgenic Nicotiana tabacum. 3 Biotech. 2018;8(6):277.
- Gao H, Wang Y, Xu P, Zhang Z. Overexpression of a WRKY transcription factor TaWRKY2 enhances drought stress tolerance in transgenic wheat. Frontiers in plant science. 2018;9:997.
- Wang Y, Jiang L, Chen J, Tao L, An Y, Cai H, et al. Overexpression of the alfalfa WRKY11 gene enhances salt tolerance in soybean. PLoS One. 2018;13(2):e0192382.
- Hu CA, Delauney AJ, Verma DP. A bifunctional enzyme (delta 1-pyrroline-5-carboxylate synthetase) catalyzes the first two steps in proline biosynthesis in plants. Proceedings of the National Academy of Sciences. 1992;89(19):9354-8.
- Hajrah NH, Obaid AY, Atef A, Ramadan AM, Arasappan D, Nelson CA, et al. Transcriptomic analysis of salt stress responsive genes in Rhazya stricta. PloS one. 2017;12(5):e0177589.
- Knauss S, Rohrmeier T, Lehle L. The auxin-induced maize gene ZmSAUR2 encodes a short-lived nuclear protein expressed in elongating tissues. Journal of Biological Chemistry. 2003;278(26):23936-43.
- Paponov IA, Paponov M, Teale W, Menges M, Chakrabortee S, Murray JAH, et al. Comprehensive transcriptome analysis of auxin responses in Arabidopsis. Molecular Plant. 2008;1(2):321-37.
- Basu S, Roychoudhury A. Expression profiling of abiotic stress-inducible genes in response to multiple stresses in rice (Oryza sativa L.) varieties with contrasting level of stress tolerance. BioMed research international. 2014;2014:706890.
- Yu Y, Wang L, Chen J, Liu Z, Park C-M, Xiang F. WRKY71 acts antagonistically against salt-delayed flowering in Arabidopsis thaliana. Plant and Cell Physiology. 2018;59(2):414-22.
- Chu X, Wang C, Chen X, Lu W, Li H, Wang X, et al. The cotton WRKY gene GhWRKY41 positively regulates salt and drought stress tolerance in transgenic Nicotiana benthamiana. PLoS One. 2015;10(11):e0143022.
- Ding ZJ, Yan JY, Li CX, Li GX, Wu YR, Zheng SJ. Transcription factor WRKY 46 modulates the development of Arabidopsis lateral roots in osmotic/salt stress conditions via regulation of ABA signaling and auxin homeostasis. The Plant Journal. 2015;84(1):56-69.
- Gao S, Zhang YL, Yang L, Song JB, Yang ZM. AtMYB20 is negatively involved in plant adaptive response to drought stress. Plant and soil. 2014;376(1-2):433-43.
- Luo D-l, Ba L-j, Shan W, Kuang J-f, Lu W-j, Chen J-y. Involvement of WRKY transcription factors in abscisic-acid-induced cold tolerance of banana fruit. Journal of agricultural and food chemistry. 2017;65(18):3627-35.
- Wang J, An C, Guo H, Yang X, Chen J, Zong J, et al. Physiological and transcriptomic analyses reveal the mechanisms underlying the salt tolerance of Zoysia japonica Steud. BMC plant biology. 2020;20(1):1-16.
- Gong Q, Li S, Zheng Y, Duan H, Xiao F, Zhuang Y, et al. SUMOylation of MYB30 enhances salt tolerance by elevating alternative respiration via transcriptionally upregulating AOX1a in Arabidopsis. The Plant Journal. 2020;102(6):1157-71.
- Zheng S, Tasnif Y, Hebert MF, Davis CL, Shitara Y, Calamia JC, et al. Measurement and compartmental modeling of the effect of CYP3A5 gene variation on systemic and intrarenal tacrolimus disposition. Clinical Pharmacology & Therapeutics. 2012;92(6):737-45.
- Hay RT. SUMO: a history of modification. Molecular cell. 2005;18(1):1-12.
- Liu L, Zhang J, Adrian J, Gissot L, Coupland G, Yu D, et al. Elevated levels of MYB30 in the phloem accelerate flowering in Arabidopsis through the regulation of FLOWERING LOCUS T. PloS one. 2014;9(2):e89799.
- Raffaele S, Vailleau F, Léger A, Joubès J, Miersch O, Huard C, et al. A MYB transcription factor regulates very-long-chain fatty acid biosynthesis for activation of the hypersensitive cell death response in Arabidopsis. The Plant Cell. 2008;20(3):752-67.
- Li L, Yu X, Thompson A, Guo M, Yoshida S, Asami T, et al. Arabidopsis MYB30 is a direct target of BES1 and cooperates with BES1 to regulate brassinosteroid‐induced gene expression. The Plant Journal. 2009;58(2):275-86.
- Ashraf M, Akram NA, Arteca RN, Foolad MR. The physiological, biochemical and molecular roles of brassinosteroids and salicylic acid in plant processes and salt tolerance. Critical Reviews in Plant Sciences. 2010;29(3):162-90.
- Jaradat MR, Feurtado JA, Huang D, Lu Y, Cutler AJ. Multiple roles of the transcription factor AtMYBR1/AtMYB44 in ABA signaling, stress responses, and leaf senescence. BMC Plant Biology. 2013;13(1):192.
- Nguyen XC, Hoang MHT, Kim HS, Lee K, Liu X-M, Kim SH, et al. Phosphorylation of the transcriptional regulator MYB44 by mitogen activated protein kinase regulates Arabidopsis seed germination. Biochemical and biophysical research communications. 2012;423(4):703-8.
- Jung C, Seo JS, Han SW, Koo YJ, Kim CH, Song SI, et al. Overexpression of AtMYB44 enhances stomatal closure to confer abiotic stress tolerance in transgenic Arabidopsis. Plant physiology. 2008;146(2):623-35.
- Devaiah BN, Madhuvanthi R, Karthikeyan AS, Raghothama KG. Phosphate starvation responses and gibberellic acid biosynthesis are regulated by the MYB62 transcription factor in Arabidopsis. Molecular plant. 2009;2(1):43-58.
- Haga N, Kobayashi K, Suzuki T, Maeo K, Kubo M, Ohtani M, et al. Mutations in MYB3R1 and MYB3R4 cause pleiotropic developmental defects and preferential down-regulation of multiple G2/M-specific genes in Arabidopsis. Plant Physiology. 2011;157(2):706-17.
- Su C-F, Wang Y-C, Hsieh T-H, Lu C-A, Tseng T-H, Yu S-M. A novel MYBS3-dependent pathway confers cold tolerance in rice. Plant physiology. 2010;153(1):145-58.
- Wang T, Tohge T, Ivakov A, Mueller-Roeber B, Fernie AR, Mutwil M, et al. Salt-related MYB1 coordinates abscisic acid biosynthesis and signaling during salt stress in Arabidopsis. Plant Physiology. 2015;169(2):1027-41.
- Cominelli E, Galbiati M, Vavasseur A, Conti L, Sala T, Vuylsteke M, et al. A guard-cell-specific MYB transcription factor regulates stomatal movements and plant drought tolerance. Current biology. 2005;15(13):1196-200.
- Kim JH, Nguyen NH, Jeong CY, Nguyen NT, Hong S-W, Lee H. Loss of the R2R3 MYB, AtMyb73, causes hyper-induction of the SOS1 and SOS3 genes in response to high salinity in Arabidopsis. Journal of Plant Physiology. 2013;170(16):1461-5.
(Visited 616 times, 1 visits today)

This work is licensed under a Creative Commons Attribution 4.0 International License.






