Manuscript accepted on : 02-04-2024
Published online on: 16-04-2024
Plagiarism Check: Yes
Reviewed by: Dr Sunil Chaudhry
Second Review by: Dr. Snehal Chakorkar
Final Approval by: Dr. Eugene A. Silow
Virtual High Throughput Screening of Marine Bacterial Metabolites for BACE1 Inhibition
Annu Grewal , Deepak Sheokand
, Deepak Sheokand , Raveena Chauhan
, Raveena Chauhan , Vandana Saini
, Vandana Saini and Ajit Kumar*
and Ajit Kumar*
Toxicology and Computational Biology Group, Centre for Bioinformatics, Maharshi Dayanand University, Rohtak, Haryana, India.
Corresponding Author E-mail: akumar.cbt.mdu@gmail.com
ABSTRACT: Alzheimer's disease is a neurodegenerative ailment reportedly caused by abnormal production or collection of amyloid-β peptides. Alzheimer-causing Aβ peptides are produced when amyloid precursor protein is cleaved by β-secretase-1 (BACE1). Previous failures in clinical trials of BACE1 inhibitors have invited studies with lesser side effects and better therapeutic efficacy. Marine bacterial metabolites have been used successfully as therapeutic options for many diseases and hence will be suitable to study for their potential as Alzheimer's disease therapeutics. The present work attempted to virtually screen marine bacterial metabolites by molecular docking studies against BACE1. A total of 2884 marine bacterial metabolites were retrieved from the Comprehensive Marine Natural Products Database and analyzed for drug-like properties, using Lipinski’s rules, ADMET profiling and binding affinities. Atabecestat was selected as the standard BACE1 inhibitor for our study. The initial screening using Lipinski’s rule selected 1357 compounds and further filtration using ADMET properties calculated 199 metabolites. Molecular docking studies against BACE1 resulted in 8 marine microbial metabolites echoside D (-9.9 kcal/mol), urdamycin N6 (-9.9 kcal/mol), echoside A (-9.7 kcal/mol), nocatrione A (-9.6 kcal/mol), nocatrione B (-9.5 kcal/mol), homoseongomycin (-9.4 kcal/mol), echoside B (-9.2 kcal/mol) and thioquinomycin A (-9.2 kcal/mol) having binding affinity higher than Atabecestat (-8.9kcal/mol).
KEYWORDS: Alzheimer’s Disease; BACE1; Marine Bacterial Compounds; Molecular Docking Studies
| Copy the following to cite this article: Grewal A, Sheokand D, Chauhan R, Saini V, Kumar A. Virtual High Throughput Screening of Marine Bacterial Metabolites for BACE1 Inhibition. Biotech Res Asia 2024;21(2). |
| Copy the following to cite this URL: Grewal A, Sheokand D, Chauhan R, Saini V, Kumar A. Virtual High Throughput Screening of Marine Bacterial Metabolites for BACE1 Inhibition. Biotech Res Asia 2024;21(2). Available from: https://bit.ly/3TYc5qn |
Introduction
Alzheimer’s disease (AD) is a gradually developing neurodegenerative disorder that interferes with memory, cognition, and behaviour. It accounts for 60-80% of all cases of dementia in older adults1. AD is characterized by the accumulation of abnormal protein deposits in the brain, called amyloid-beta (Aβ) plaques and neurofibrillary tangles (NFTs). Aβ plaques are clumps of protein fragments that build up between neurons and hinder their communication. NFTs are twisted fibers of tau protein, that form inside neurons and disrupt their function and structure. These protein deposits impair the function and survival of neurons, leading to brain atrophy and cognitive decline. The clinical manifestations of AD typically begin with mild memory impairment and executive dysfunction and progressively deteriorate over time, resulting in confusion, disorientation, aphasia, mood disorders, and behavioural disturbances. There is no definitive treatment, but pharmacological interventions can help delay the progression of the disease1.
BACE1 is a transmembrane aspartyl protease with 2 aspartate molecules (Asp93 and Asp289) as the active site residues2, responsible for the catalysis of the amyloidogenic pathway in AD, by cleaving amyloid precursor protein (APP) at the β-site and releasing the N-terminal fragment of Aβ. BACE1 expression and activity are elevated in AD brains, which may contribute to the increased production and aggregation of Aβ3. BACE1 is expressed in neurons, astrocytes, oligodendrocytes, and endothelial cells, and its activity is modulated by several factors, such as pH, oxidative stress, calcium levels, and inflammation4. BACE1 is not only involved in Aβ generation, but also in the processing of other substrates that have important roles in synaptic function, myelination, and neuronal survival5. Recent setbacks in clinical trials of BACE1 inhibitors have invited studies with lesser side effects and better therapeutic efficacy.
Marine bacterial metabolites have gained attention in recent years for their potential as a source of new therapeutic options. They have been found to exhibit a wide range of biological activities, including antimicrobial, anti-inflammatory, anticancer, cardio and neuroprotective properties6. The exploration of marine bacterial metabolites as therapeutics offers the advantage of causing fewer adverse effects compared to synthetic drugs. Marine bacterial metabolites have been successfully used as therapeutic options for AD7,8 and have reported BACE1 inhibitory properties9. The present work attempted to virtually screen marine bacterial metabolites from the Comprehensive Marine Natural Products Database10 by molecular docking studies against BACE1.
Materials and Methodology
Retrieval of marine bacterial metabolites
Marine bacterial metabolites were retrieved from the Comprehensive Marine Natural Products Database (CMNPD) on 02/02/2023. Presently, CMNPD has more than 30,000 entries of marine natural products with various pharmacokinetic properties10. Taxonomy was chosen as “Bacteria” and the data of structural and physiological parameters was retrieved in .sdf and .tsv file formats respectively.
Lipinski’s rule of five and ADMET profiling
The retrieved marine bacterial compounds were screened for Lipinski’s rule of five based on the physiochemical properties. According to Lipinski’s rule of five, “an orally active drug can have no more than one violation of these conditions: No more than 5 hydrogen bond donors, No more than 10 hydrogen bond acceptors, Molecular mass less than 500 Da, Partition coefficient not greater than 5”. ADMET (Absorption, Distribution, Metabolism, Excretion and Toxicity) profiling was done based on Blood-brain barrier (BBB) permeability, Human intestinal absorption (HIA), Caco-2, Drug-induced liver injury (DILI), Human Hepatotoxicity (H-HT), Ames toxicity, Respiratory toxicity, Carcinogenicity and human ether-à-go-go related gene (hERG) using ADMETlab2.011. ADMETlab2.0 is an open-access tool for the prediction of physicochemical properties in medicinal chemistry. The compounds obtained after ADMET screening were selected for further studies.
Retrieval and optimisation of the receptor
Retrieval of the selected receptor protein (BACE1) for virtual screening was done using the Alpha Fold database12. Alpha Fold is an open-access database for over 200 million predicted protein structures. Receptor preparation and optimization were done using Chimera13. For optimization, default parameters were used: “Gasteiger” charges were added and “AMBER ff99bsc0” was used as a forcefield parameter, followed by energy minimization using the “Steepest decent (100 steps, with a step size of 0.02 Angstrom)” and “Conjugate gradient (10 steps, with a step size of 0.02)”. Atabecestat (PubChem CID:68254185) was selected as the standard drug for the inhibition studies of BACE15.
Molecular docking studies
DockPrep of selected compounds obtained after Lipinski’s rule of five and ADMET profiling was done along with their energy minimization using OpenBabel14 with Pyrx15 and molecular docking studies were performed using AutoDock Vina16 with Pyrx. Pyrx is an open-access software for the virtual screening of small molecules library. BACE1 was selected as a receptor and converted to macromolecule as .pdbqt file format. Grid box parameters were set to maximum (Center X: -5.9446 Y: 3.0334 Z: -2.9837, Dimensions (Angstrom) X: 67.3296 Y: 53.0806 Z: 60.4251) and blind docking was performed at exhaustiveness of 8. The compounds having binding affinity higher than Atabecestat were selected for further studies of binding pocket analysis.
Binding pocket analysis
Binding pocket analysis of the docked complexes of the selected marine bacterial compounds against BACE1 was done using LigPlot+17. LigPlot+ automatically generates 2D diagrams of interacting residues from 3D coordinates of the ligand-receptor complex.
Result and discussions
Retrieval of marine bacterial metabolites
A total of 2884 marine bacterial metabolites were retrieved from the Comprehensive Marine Natural Products Database (CMNPD) on 02/02/2023. All the compounds had specific database identifiers (CMNPD COMPOUND_ID) and the file downloaded in .sdf format had specific structural information for all the compounds. The file downloaded in .tsv format had information about various physiochemical parameters (Supplementary 1). The physiochemical parameter file was used for the screening of the marine bacterial compounds against Lipinski’s rule and ADMET profiling. The structural files of the bacterial metabolites were used for the molecular docking studies.
Lipinski’s rule and ADMET profiling
The .tsv file of retrieved compounds was screened for Lipinski’s rule (HBA, HBD, ALOGP, and MOLECULAR_MASS) and it was found that a total of 1357 followed the rule of five (Supplementary 2). ADMET (Absorption, Distribution, Metabolism, Excretion and Toxicity) profiling was done based on the excellent empirical decision of the ADMETlab2.0 tool. BBB permeability, HIA, Caco-2, DILI, H-HT, Ames toxicity, Respiratory toxicity, Carcinogenicity and hERG empirical evaluation resulted in 199 compounds (Supplementary 3).
Retrieval and optimisation of the receptor
BACE1 was retrieved from the Alpha Fold database with ID: AF-P56817-F1. The color scheme represented the model confidence, known as the per-residue model confidence score (pLDDT) score (Figure 1) and it was found that the major domains (blue color) of the receptor structure were falling under a very high confidence score (pLDDT > 90). The predicted aligned error is represented by a graph between aligned residues and the expected position error in Angstroms (Figure 2), and maximum residues were found to be aligned at the correct position (dark green lines). Based on the pLDDT score, the receptor portion falling under the low score (from sequence lengths 1-55 and 448-501) was truncated, leading to a high-confidence model with 56 to 447 amino acids (Figure 3). The truncated structure of the selected target receptor protein BACE1(from sequence length 56-447) was optimised using Chimera and saved in .pdb file format for molecular docking studies.
 |
Figure 1: Complete structure of BACE1 retrieved from Alpha fold database (AF-P56817-F1). Alpha Fold provides a per-residue confidence score (pLDDT) between 0 to 100. |
 |
Figure 2: The predicted aligned error of the receptor structure, where the dark green area represents good confidence and low error whereas the light green area represents no confidence and high error. |
 |
Figure 3: The sequence of the prepared receptor structure (The yellow blocks represent alpha-helix and the green blocks represent beta-strands in the receptor structure). |
Molecular docking studies
Molecular docking studies of the selected 199 ligand compounds were performed against the prepared truncated receptor protein BACE1 (Supplementary 4). The resulted output files of the docking studies were analysed for their binding poses and binding energies at different root mean square deviations (RMSD). The binding energy was selected for the binding pose having a RMSD value of zero. Standard drug compound (Atabecestat) had binding energy of -8.9 kcal/mol at RMSD value of zero and it was found that 8 ligand compounds CMNPD24462 (-9.9 kcal/mol), CMNPD30021 (-9.9 kcal/mol), CMNPD24459 (-9.7 kcal/mol), CMNPD24408 (-9.6 kcal/mol), CMNPD24409 (-9.5 kcal/mol), CMNPD23207 (-9.4 kcal/mol), CMNPD24460 (-9.2 kcal/mol) and CMNPD30040 (-9.2 kcal/mol) had binding energy lower than Atabecestat (Table1). 4 metabolites- echoside A (CMNPD24459), echoside B (CMNPD24460), echoside D (CMNPD24462) and thioquinomycin A (CMNPD30040) belongs to Streptomyces marinus while 2 metabolites- nocatrione A (CMNPD24408) and nocatrione B (CMNPD24409) are from Nocardiopsis aegyptia while urdamycin N6 (CMNPD30021) and homoseongomycin (CMNPD23207) are the metabolites of Streptomyces diastaticus and Salinispora pacifica, respectively, as reported in the CMNPD database.
Binding pocket analysis
The selected 8 CMNPD compounds Echoside D (CMNPD24462), Urdamycin N6 (CMNPD30021), Echoside A (CMNPD24459), Nocatrione A (CMNPD24408), Nocatrione B (CMNPD24409), Homoseongomycin (CMNPD23207), Echoside B (CMNPD24460) and Thioquinomycin A (CMNPD30040), along with the reference drug selected for the study (Atabecestat) were analysed for the binding pocket interactions with BACE1 using LigPlot+. Active-site inhibition of the BACE1 was observed against Atabecestat, involving H-bonding and other non-covalent (hydrophobic) interactions, inside the binding pocket of BACE1. The active site residues of BACE1 (Asp93 and Asp289) were observed to participate in the binding pocket interactions of the Atabecestat with BACE1. Atabecestat showed interaction with 9 residues, involving one H-bond with Thr133 and other non-covalent interactions with Asp93, Tyr132, Phe169, Tyr259, Lys285, Ile287, Gly291 and Thr390 (Figure 4, Table 2).
Table 1: Marine bacterial metabolites of the CMNPD database having binding affinity, for BACE1, better than the standard (Atabecestat) selected for the study.
| S.No. | PubChem ID | CMNPD ID | Binding energy (kcal/mol) | Name of compound | Microbial source |
| 1. | 76310781 | CMNPD24462 | -9.9 | Echoside D | Streptomyces marinus |
| 2. | 139590640 | CMNPD30021 | -9.9 | Urdamycin N6 | Streptomyces diastaticus |
| 3. | 76332530 | CMNPD24459 | -9.7 | Echoside A | Streptomyces marinus |
| 4. | 101888872 | CMNPD24408 | -9.6 | Nocatrione A | Nocardiopsis aegyptia |
| 5. | 101888873 | CMNPD24409 | -9.5 | Nocatrione B | Nocardiopsis aegyptia |
| 6. | 139587456 | CMNPD23207 | -9.4 | Homoseongomycin | Salinispora pacifica |
| 7. | 45782902 | CMNPD24460 | -9.2 | Echoside B | Streptomyces marinus |
| 8. | 139590302 | CMNPD30040 | -9.2 | Thioquinomycin A | Streptomyces marinus |
| 9. | 68254185 | Standard | -8.9 | Atabecestat | NA |
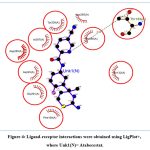 |
Figure 4: Ligand-receptor interactions were obtained using LigPlot+, where Unk1(N)= Atabecestat. |
All the selected compounds were observed to share a similar binding pocket as Atabecestat. CMNPD23207 formed 5 H-bonds with Thr133, Gly291, Thr293, Arg296 and other similar non-covalent interactions with Asp93, Tyr132, Phe169, Tyr259 and Ile287 (Figure 5a, Table 2). Similarly, CMNPD24408 showed 2 H-bonding with Thr133 and Leu324, while similar non-covalent interactions with Tyr132, Gly291 and Thr292 (Figure 5b, Table 2). Comparably, CMNPD24409 formed 3 H-bonds with Thr292 and Arg368, along with other common interactions involving Tyr132, Phe169 and Gly291 (Figure 5c, Table 2). CMNPD24459 formed a maximum of 8 H-bonds with Asp93, Gly95, Thr133, Asp289, Thr292 and Arg296, in addition to similar non-covalent interactions with Tyr132, Phe169, Tyr259, Ile287 and Gly291 (Figure 5d, Table 2). CMNPD24460 showed 6 H-bonding interactions with Gly95, Thr133, Asp289 and Thr292, and shared non-covalent interactions with Tyr132, Phe169 and Gly291 (Figure 5e, Table 2). CMNPD24462 showed 4 H-bonding with 3 residues Thr133, Thr292 and Arg296, while a minimum of 2 common interactions with Tyr132 and Phe169 (Figure 5f, Table 2). CMNPD30021 formed 4 H-bonds with Lys168, Thr293, Asn294 and Arg296, and other non-covalent interactions with Tyr132 and Phe169 (Figure 5g, Table 2). CMNPD30040 showed 2 H-bonds with Thr133 and Tyr259 and Tyr132 and Thr390 as similar binding pocket residues (Figure 5h, Table 2).
Looking into our findings and reported studies, the selected metabolites from Streptomyces sp., Nocardiopsis sp. and Salinispora sp. can be considered for BACE1 inhibitory therapeutics design. Literature studies have suggested that Streptomyces sp. strains have reported therapeutic activities for acetylcholinesterase and SARS-CoV-218, 19, therefore the selected metabolites from Streptomyces marinus and Streptomyces diastaticus (echoside A, echoside B, echoside D, thioquinomycin A and urdamycin N6) can be considered for BACE1 inhibitory therapeutics design. Nocardiopsis sp. metabolites have reported antiphotoaging therapeutic activity20, inferring the therapeutic potential of selected metabolites (nocatrione A, nocatrione B). Homoseongomycin has reported therapeutic potential against encephalitic alphaviruses21. The secondary metabolites produced by Streptomyces sp. have been shown to act as dual inhibitors of BACE1 and Aβ aggregation22. Tirandamycin B isolated from a newly found Streptomyces composti sp. nov. have also been shown to have the neuroprotective and beta-secretase1 inhibitor profiles23.
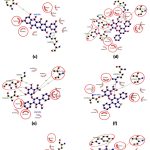 |
Figure 5 (a-h): Ligand-receptor interactions obtained using Ligplot+. where (a): CMNPD23207, (b): CMNPD24408, (c): CMNPD24409, (d): CMNPD24459, (e): CMNPD24460, (f): CMNPD24462, (g): CMNPD30021, (h): CMNPD30040. |
Table 2: Interacting residues of BACE1 when docked against marine bacterial metabolites of CMNPD database with binding score better than the standard (Atabecestat).
| S. No. | Name | No of H-bonds | H-bond forming residues | Other interacting residues |
| 1. | Atabecestat | 1 | Thr133 (3.02Å) | Asp93, Tyr132, Phe169, Tyr259, Lys285, Ile287, Gly291, Thr390 |
| 2. | CMNPD23207 | 5 | Thr133 (2.98Å and 2.80Å), Gly291 (2.94Å), Thr293 (3.07Å), Arg296 (2.80Å) | Asp93, Tyr132, Phe169, Ile171, Trp176, Ile179, Tyr259, Ile287, Asp289 |
| 3. | CMNPD24408 | 2 | Thr133 (3.23Å), Leu324 (2.73Å) | Tyr132, Gln134, Asp289, Gly291, Thr292, Thr293, Asn294, Arg296, Gly325 |
| 4. | CMNPD24409 | 3 | Thr292 (3.32Å), Arg368 (2.83Å and 3.25Å) | Tyr132, Phe169, Ile171, Ile179, Gly291, Thr293, Asn294, Gly325 |
| 5. | CMNPD24459 | 8 | Asp93 (3.25Å), Gly95 (3.04Å and 2.81Å), Thr133 (2.85Å), Asp289 (3.24Å and 3.05Å), Thr292 (2.70Å)
Arg296 (2.97Å). |
Tyr132, Gln134, Phe169, Ile179, Tyr259, Ile287, Gly291, Asn294 |
| 6. | CMNPD24460 | 6 | Gly95 (3.26Å), Thr133 (2.84Å and 2.88Å), Asp289 (2.70Å and 2.91Å)
Thr292 (2.74Å) |
Tyr132, Gln134, Phe169, Ile179, Gly291, Thr293, Asp294, Arg296, Ser386 |
| 7. | CMNPD24462 | 4 | Thr133 (3.09Å), Thr292 (2.82Å)
Arg296 (2.95Å and 3.12Å) |
Tyr132, Gln134, Phe169, Ile179, Gly195, Asp294, Ser386, Val393 |
| 8. | CMNPD30021 | 4 | Lys168 (2.89Å), Thr293 (3.07Å), Asn294 (2.83Å), Arg296 (2.93Å) | Tyr132, Thr133, Gln134, Gly135, Phe169, Ile171, Leu324, Ser386 |
| 9. | CMNPD30040 | 2 | Thr133 (3.11Å), Tyr259 (2.82Å) | Leu91, Pro131, Tyr132, Gln134, Ile171, Trp176, Thr390, Val393 |
Conclusion
The present investigation of evaluating marine bacterial metabolites of CMNPD revealed that 08 out of 2884 studied marine bacterial metabolites, showed better binding affinities as compared to the standard compound – Atabecestat, when docked against BACE-1, the target therapeutic receptor for AD. These compounds Echoside D (CMNPD24462), Urdamycin N6 (CMNPD30021), Echoside A (CMNPD24459), Nocatrione A (CMNPD24408), Nocatrione B (CMNPD24409), Homoseongomycin (CMNPD23207), Echoside B (CMNPD24460) and Thioquinomycin A (CMNPD30040), can be worth evaluating as the potential BACE1 inhibitors for AD therapeutics inviting further in-vitro and in-vivo validation.
Acknowledgements
The authors thank UGC, New Delhi for providing SJSGC-JRF to AG and M. D. University, Rohtak for providing URS to DS. The authors also thank DBT-BUILDER MDU for providing laboratory facility for the work.
Conflict of interest
There are no conflict of interest
Funding Sources
There is no funding sources
Ethics Approval Statement
The work does not involve any experiment on human or animal.
References
- 2022 Alzheimer’s disease facts and figures. Alzheimers Dement. 2022;18(4):700-789. doi:10.1002/alz.12638
CrossRef - Cole SL, Vassar R. The Alzheimer’s disease beta-secretase enzyme, BACE1. Mol Neurodegener. 2007;2:22. Published 2007 Nov 15. doi:10.1186/1750-1326-2-22
CrossRef - Hampel H, Lista S, Vanmechelen E, et al. β-Secretase1 biological markers for Alzheimer’s disease: state-of-art of validation and qualification. Alzheimers Res Ther. 2020;12(1):130. Published 2020 Oct 16. doi:10.1186/s13195-020-00686-3
CrossRef - Yan R. Physiological Functions of the β-Site Amyloid Precursor Protein Cleaving Enzyme 1 and 2. Front Mol Neurosci. 2017;10:97. Published 2017 Apr 19. doi:10.3389/fnmol.2017.00097
CrossRef - Vassar R. BACE1 inhibitor drugs in clinical trials for Alzheimer’s disease. Alzheimers Res Ther. 2014;6(9):89. Published 2014 Dec 24. doi:10.1186/s13195-014-0089-7
CrossRef - Abdelghani Z, Hourani N, Zaidan Z, Dbaibo G, Mrad M, Hage-Sleiman R. Therapeutic applications and biological activities of bacterial bioactive extracts. Arch Microbiol. 2021;203(8):4755-4776. doi:10.1007/s00203-021-02505-1
CrossRef - Zhu B, Li Z, Qian PY, Herrup K. Marine bacterial extracts as a new rich source of drugs against Alzheimer’s disease. J Neurochem. 2020;152(4):493-508. doi:10.1111/jnc.14847
CrossRef - Rathnayake AU, Abuine R, Kim YJ, Byun HG. Anti-Alzheimer’s Materials Isolated from Marine Bio-resources: A Review. Curr Alzheimer Res. 2019;16(10):895-906. doi:10.2174/1567205016666191024144044
CrossRef - Williams P, Sorribas A, Liang Z. New methods to explore marine resources for Alzheimer’s therapeutics. Curr Alzheimer Res. 2010;7(3):210-213. doi:10.2174/156720510791050812
CrossRef - Lyu C, Chen T, Qiang B, et al. CMNPD: a comprehensive marine natural products database towards facilitating drug discovery from the ocean. Nucleic Acids Res. 2021;49(D1):D509-D515. doi:10.1093/nar/gkaa763
CrossRef - Xiong G, Wu Z, Yi J, et al. ADMETlab 2.0: an integrated online platform for accurate and comprehensive predictions of ADMET properties. Nucleic Acids Res. 2021;49(W1):W5-W14. doi:10.1093/nar/gkab255
CrossRef - Varadi M, Anyango S, Deshpande M, et al. AlphaFold Protein Structure Database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2022;50(D1):D439-D444. doi:10.1093/nar/gkab1061
CrossRef - Pettersen EF, Goddard TD, Huang CC, et al. UCSF Chimera–a visualization system for exploratory research and analysis. J Comput Chem. 2004;25(13):1605-1612. doi:10.1002/jcc.20084
CrossRef - O’Boyle NM, Banck M, James CA, Morley C, Vandermeersch T, Hutchison GR. Open Babel: An open chemical toolbox. J Cheminform. 2011;3:33. Published 2011 Oct 7. doi:10.1186/1758-2946-3-33
CrossRef - Dallakyan S, Olson AJ. Small-molecule library screening by docking with PyRx. Methods Mol Biol. 2015;1263:243-250. doi:10.1007/978-1-4939-2269-7_19
CrossRef - Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31(2):455-461. doi:10.1002/jcc.21334
CrossRef - Laskowski RA, Swindells MB. LigPlot+: multiple ligand-protein interaction diagrams for drug discovery. J Chem Inf Model. 2011;51(10):2778-2786. doi:10.1021/ci200227u
CrossRef - Almasi F, Mohammadipanah F, Adhami HR, Hamedi J. Introduction of marine-derived Streptomyces sp. UTMC 1334 as a source of pyrrole derivatives with anti-acetylcholinesterase activity. J Appl Microbiol. 2018;125(5):1370-1382. doi:10.1111/jam.14043
CrossRef - Melinda YN, Widada J, Wahyuningsih TD, Febriansah R, Damayanti E, Mustofa M. Metabologenomics approach to the discovery of novel compounds from Streptomyces sp. GMR22 as anti-SARS-CoV-2 drugs. Heliyon. 2021;7(11):e08308. doi:10.1016/j.heliyon.2021.e08308
CrossRef - Kim MC, Hwang E, Kim T, Ham J, Kim SY, Kwon HC. Nocatriones A and B, photoprotective tetracenediones from a marine-derived Nocardiopsis sp. J Nat Prod. 2014;77(10):2326-2330. doi:10.1021/np5006086
CrossRef - Lin SC, Lehman CW, Stewart AK, et al. Homoseongomycin, a compound isolated from marine actinomycete bacteria K3-1, is a potent inhibitor of encephalitic alphaviruses. Antiviral Res. 2021;191:105087. doi:10.1016/j.antiviral.2021.105087
CrossRef - Yokoya M, Nakai K, Kawashima M, et al. Inhibition of BACE1 and amyloid β aggregation by polyketide from Streptomyces sp. Chem Biol Drug Des. 2022;99(2):264-276. doi:10.1111/cbdd.13980
CrossRef - Duangupama T, Pratuangdejkul J, Chongruchiroj S, et al. New insights into the neuroprotective and beta-secretase1 inhibitor profiles of tirandamycin B isolated from a newly found Streptomyces composti sp. nov. Sci Rep. 2023;13(1):4825. Published 2023 Mar 24. doi:10.1038/s41598-023-32043-3
CrossRef

This work is licensed under a Creative Commons Attribution 4.0 International License.





