Manuscript accepted on : 15-03-2024
Published online on: 25-03-2024
Plagiarism Check: Yes
Reviewed by: Dr. Neha Jamwal
Second Review by: Dr. Eric M. Hallerman
Final Approval by: Dr Amr Salah Morsy Amine Selem
Chaueichongla Phom1 and Jeyaparvarthi Somasundaram2*
and Jeyaparvarthi Somasundaram2*
Department of Zoology, St. Joseph University, Dimapur, India
corresponding Author E-mail:jeya5001@gmail.com
DOI : http://dx.doi.org/10.13005/bbra/3199
ABSTRACT: DNA barcoding is a method of species identification that revolutionized the way we study and understand biodiversity. With advancements in molecular genetics and DNA sequence archives, it has become possible to use short DNA sequences to identify species, even when the specimens are difficult to distinguish by traditional morphological methods. In this article, we explore the utility of DNA barcoding in the Tuensang ecosystem of Nagaland, India and evaluate its effectiveness for species identification, informing ongoing conservation of populations and species. A total of 62 species, which included insects, spiders, lizards, birds and mammals, were collected from Tuensang areas of northeast India and identified using DNA barcodes. DNA was extracted from muscle tissue and PCR was done with two pairs of primers targeting the mitochondrial COI gene. Sanger sequencing was employed and the obtained sequences were analysed to identify the species and reconstruct the evolutionary relationships amongst them. Our results provided molecular characterization of species from Tuensang areas of Nagaland for the first time.
KEYWORDS: Biological resources; COI; DNA barcoding; Evolutionary relationship; Extinction; Sanger sequencing
Download this article as:| Copy the following to cite this article: Phom C, Somasundaram J. Molecular Identification of the Diversity of Insects, Spiders, Lizards, Birds and Mammals of Tuensang District, Nagaland, India. Biotech Res Asia 2024;21(1). |
| Copy the following to cite this URL: Phom C, Somasundaram J. Molecular Identification of the Diversity of Insects, Spiders, Lizards, Birds and Mammals of Tuensang District, Nagaland, India. Biotech Res Asia 2024;21(1). Available from: https://bit.ly/3PBP2Ar |
Introduction
“Biodiversity” refers to the variety of living organisms from all sources, including terrestrial, marine, and other aquatic ecosystems as well as the ecological complexes to which they belong. At the same time, the presence of various faunal genetic resources located in any region or country is a component of animal biodiversity. Genetic diversity is the basis for a population to evolve and adapt to rapid changes in the environment25. Today’s biodiversity provides the opportunity for sustaining vital environmental services that support life on Earth. Therefore, it is crucial to prioritise species protection and sustainable use. The identification of species paves the way to a deeper understanding of evolutionary processes among varied animal lineages by recording information on patterning and the range of variation.
Various methods have been developed over the decades for the characterization of wild animals which are largely categorized into phenotypic, biochemical and molecular markers. The history of taxonomy and evolutionary research is rooted in the study of morphology. For many years, the classification of species has been most heavily influenced by morphology in its widest meaning, which includes all expressions of structure and form. Morphology was used as a fundamental organising principle to arrange an apparently chaotic variety of living forms into higher (“macrotaxonomic”) levels initially17. Recently, the DNA barcoding approach based on the cytochrome c oxidase subunit I (COI) gene present in mitochondrial DNA has become adopted as a global biological identification method for all species due to its accuracy when compared to existing classical taxonomic methods21. This approach uses DNA extraction, sequencing and barcoding for genomic characterisation8, and it has become as an efficient and reliable tool for identifying, confirming and resolving closely related taxa19. Molecular techniques are increasingly employed in conservation of biological diversity in response to the alarming extinction of different species. Therefore, many conservation geneticists study genetic markers to make decisions for conservation of endangered wild animals. Here, choosing of effective profiling technique is a very critical step, as incorrect data on molecular information may result in incorrect conservation actions29. In this regard, the application of both karyotypes and molecular genetic markers to conserve wild animals has been put into practice24. Identification and characterization of species are two important steps towards crafting sustainable conservation strategies for wild animals. Studying genetic diversity within and between populations, estimation of effective population size and assessment of possible bottlenecks are practices carried out for characterization of wild animal populations.
Molecular characterization is based on studies of both mitochondrial DNA (mtDNA) and nuclear DNA1. In recent years, advances in molecular techniques have encouraged the application of simple and precise DNA analysis in the taxonomic field. Among the existing genome-based approaches, DNA barcoding stands as a robust strategy to identify existing species and to discover unknown species through comparative analysis of sequence variation2,18. Extensive research has shown that DNA barcoding can identify a wide range of animal species, including mammals, reptiles, birds, fishes, amphibians and crustaceans12,28,30. The classification and identification of various life forms, particularly insects have been a major challenge to the scientific community especially with the dwindling interest and funding for taxonomy16. DNA barcoding provides an alternative approach, allowing expedited examination of species richness of all animal lineages at comparatively low cost. Its capacity to enhance progress depends on the fact that members of most species form a distinct barcode cluster7,22,23; this relationship has now been operationalized for large-scale surveys by allotting a barcode index number (BIN)9,20.
There has never been a molecular marker-based study of native faunal species of Tuensang areas of northeast India. Hence, our study aimed to characterize the diversity of animals from these areas using the DNA barcoding technique. Further, the inherent phylogenetic relationships present among those organisms were analyzed. The main objective of this study was to develop a reference library with generated barcodes.
Materials and methods
Study area
One of Nagaland’s largest and most eastern districts is Tuensang, Northeast India. The district of Tuensang covers 4,228 square kilometres and can be found at latitude of 26º 14ʹ 8.67ʺN and longitude of 94º 48ʹ 47.47ʺE, with elevations ranging from 800 to 3500m above mean sea level.
 |
Figure 1: Map of Tuensang District, Nagaland, India.
|
Species collection
For molecular characterization, animal species were collected from selected areas of Tuensang district, Nagaland, viz., Tuensang town, Tuensang village, Hakchang village, Helipong village, Ngangpong village, Sangchen compound village, Chendang village, Chingmei village and Momching village. Field research was conducted in this study area to collect the species from forest and agricultural field habitats using different techniques, such as hand collection, pitfall trap, sweep netting, snares, catapults, and air guns. The collected animal tissues were immediately stored in 100% ethanol. For carrying out this research work, a consent letter was obtained from the community leader of the district. The study was conducted from 2019 to 2022.
DNA isolation
The extraction of DNA from collected animal tissue was performed using the Qiagen Tissue Kit following the manufacturer’s protocol. 100 mg of tissue sample was mixed with 180μl of ATL buffer, 20μl of Proteinase K, vortexed, and incubated at 60°C in a dry bath for 4 hours. 200μl of AL buffer was added and mixed in, followed by a 20-minute incubation at 60°C. 200μl of ethanol was poured and put to a DNA silica column for 1 minute of spinning at 12000 rpm. 700 μl of AW1 washed twice, with the AW2 buffer spinning at 12000 rpm for 1 minute. By adding elution buffer to 20 μl of DNA, DNA was eluted and measured using the 1% agarose gel electrophoresis technique.
PCR Amplification and Gel purification
PCR amplification of the isolated DNA samples was done using universal COI primers26. The amplification reaction was performed in 25 μl reaction mixtures with DNA template of 40-80ng. The reaction mixture included 1μl of both forward and reverse primers and 25μl of the master mix containing 1μl dNTPs, 0.5μl Taq DNA polymerase (Barcode Biosciences), 5μl 10x buffer and 14.5μl distilled water. The primers were standard primers available for COI gene amplification, as below:
COIF- GGTCAACAAATCATAAAGATATTGG- Tm 510C and
COIR- TAAACTTCAGGGTGACCAAAAAATCA- Tm 530C. The PCR amplification conditions included an initial denaturation step at 94ºC for 3.5 minutes, 60ºC for 30 seconds, and 72ºC for 1 minute for 35 cycles. On a 1% agarose gel electrophoresis stained with ethidium bromide, PCR product quality was examined.
Sanger Sequencing
An Applied Biosystems 3130xl Genetic Analyzer was used to carry out bidirectional sequencing of PCR products at BioEdge Solutions, Bangalore, India. The data from the sequencing machine was collected and processed in Finch TV (https://finchtv.software.informer.com/1.4/). The obtained electropherogram files were analysed for base-calling peaks in ABI format, which was further converted to pdf and fasta files using Sequence Scanner 2 Software (https://sequence-scanner-software.software.informer.com/2.0/). The sequence data obtained during this study were subjected to NCBI-BLAST in the nucleotide database of GenBank (http://blast.ncbi.nlm.nih.gov/) to identify closely related species.
Construction of phylogenetic tree
To comprehend the link between unknown sequences and relevant top species found by NCBI-BLAST, all the sequences were aligned using the Clustal Omega software ((https://www.ebi.ac.uk/Tools/msa/clustalo/). In this manner, phylogenetic trees were created informing us of the evolutionary relationships among closely related species.
Results and Discussion
DNA barcodes provided a quick, accurate and affordable way to identify species, even those that are challenging to differentiate based on morphology or life stage. A total of 62 species, which included animals belonging to different classes, such as Insecta, Arachnida, Reptilia, Aves and Mammalia, were identified using DNA barcoding technique for the first time from the study areas of Tuensang, Nagaland. The sequences obtained from samples were submitted to GenBank, and their accession numbers and sequence lengths are reported in Table 1. Among 62 sequences, 8 COI sequences that were previously unknown were added to the GenBank database. These sequences species were Cettia brunnifrons (Hodgson, 1845), Psilopogon virens (Boddaert, 1789), Megalaima franklinii (Blyth, 1842), Turdus boulboul (Latham, 1790), Hypsipetes leucocephalus (Gmelin, JF, 1789), Sitta formosa (Blyth, 1843), Oriolus traillii (Vigors, 1832) and Hypsipetes thompsoni (Bingham, 1900). These accession numbers include: OQ920217, OQ920214, OQ920218, OQ920219, OQ780819, OQ920213, OQ920220 and OQ780820, respectively.
Table 1: Details of accession numbers and amplicon sequence length of insects, spiders, lizards, birds and mammals taken for molecular study.
|
S.no |
Common name |
Scientific name |
Family |
Accession numbers obtained for each samples |
Sequence length |
*IUCN 3.1 |
|
1 |
Red-whiskered bulbul bird |
Pycnonotus jocosus |
Pycnonotidae |
OQ874330 |
623 bp |
Least concern |
|
2 |
Indian scops owl |
Otus bakkamoena |
Strigidae |
OQ821222 |
642 bp |
Least concern |
|
3 |
Streak-throated barwing bird |
Actinodura waldeni |
Leiothrichidae |
OQ874334 |
608 bp |
Least concern |
|
4 |
Scaly thrush bird |
Zoothera dauma |
Turdidae |
OQ821223 |
639 bp |
Least concern |
|
5 |
Blyth’s tragopan |
Tragopan blythii |
Phasianidae |
OQ874335 |
655 bp |
Vulnerable |
|
6 |
Mountain bamboo partridge bird |
Bambusicola fytchii |
Phasianidae |
OQ874336 |
616 bp |
Least concern |
|
7 |
Red-faced liocichla bird |
Liocichla phoenicea |
Leiothrichidae |
OQ874338 |
609 bp |
Least concern |
|
8 |
Mrs. Gould’s sunbird |
Aethopyga gouldiae |
Nectariniidae |
OQ874339 |
632 bp |
Least concern |
|
9 |
Red-vented bulbul bird |
Pycnonotus cafer |
Pycnonotidae |
OQ821233 |
652 bp |
Least concern |
|
10 |
Beautiful nuthatch bird |
Sitta formosa |
Sittidae |
OQ920213 |
624 bp |
Vulnerable |
|
11 |
Great barbet bird |
Psilopogon virens |
Megalaimidae |
OQ920214 |
648 bp |
Least concern |
|
12 |
kalij pheasant bird |
Lophura leucomelanos |
Phasianidae |
OQ920215 |
640 bp |
Least concern |
|
13 |
Grey-sided bush warbler bird |
Cittia brunnifrons |
Cettiidae |
OQ920217 |
621 bp |
Least concern |
|
14 |
Golden-throated Barbet bird |
Magalaima franklinii |
Megalaimidae |
OQ920218 |
645 bp |
Least concern |
|
15 |
Grey-winged blackbird |
Turdus boulboul |
Turdidae |
OQ920219 |
624 bp |
Least concern |
|
16 |
Maroon oriole |
Oriolus traillii |
Oriolidae |
OQ920220 |
659 Bp |
Least concern |
|
17 |
Black bulbul bird |
Hypsipetes leucocephalus |
Pycnonotidae |
OQ780819 |
632 bp |
Least concern |
|
18 |
White-headed bulbul bird |
Hypsipetes thompsoni |
Pycnonotidae |
OQ780820 |
624 bp |
Least concern |
|
19 |
Orange-bellied leafbird |
Chloropsis hardwickii |
Chloropseidae |
OQ780821 |
654 bp |
Least concern |
|
20 |
Rufous-breasted antthrush bird |
Formicarius rufipectus |
Formicariidae |
OQ780843 |
620 bp |
Least concern |
|
21 |
Spotted linsang |
Prionodon pardicolor |
Prionodontidae |
OQ874331 |
646 bp |
Least concern |
|
22 |
Masked palm civet |
Paguma larvata |
Viverridae |
OQ874332 |
626 bp |
Least concern |
|
23 |
Southern red muntjac |
Muntiacus muntjak |
Cervidae |
OQ874333 |
623bp |
Least concern |
|
24 |
Northern flying squirrel |
Glaucomys sabrinus |
Sciuridae |
OQ821224 |
652 bp |
Least concern |
|
25 |
Swinhoe’s striped squirrel |
Tamiops swinhoei |
Sciuridae |
OQ821225 |
657 bp |
Least conncern |
|
26 |
Sambar deer |
Rusa unicolor |
Cervidae |
OQ874337 |
624 bp |
Vulnerable |
|
27 |
Himalayan striped squirrel |
Tamiops mcclellandii |
Sciuridae |
OQ920212 |
624 bp |
Least concern |
|
28 |
Orange-bellied Himalayan squirrel |
Dremomys lokriah |
Siuridae |
OQ920216 |
624 bp |
Least concern |
|
29 |
Tanezumi rat |
Rattus tanezumi |
Muridae |
OQ780822 |
623bp |
Least concern |
|
30 |
Big brown bat |
Eptesicus fuscus |
Vespertilionidae |
OQ780827 |
613 bp |
Least concern |
|
31 |
Cave myotis bat |
Myotis velifer |
Vespertilionidae |
OQ780828 |
620 bp |
Least concern |
|
32 |
Spotted forest skink |
Sphenomorphus maculatus |
Scincidae |
OQ780823 |
619 bp |
Not evaluated |
|
33 |
Farooq’s garden lizard |
Calotes farooqi |
Agamidae |
OQ780824 |
626 bp |
Not evaluated |
|
34 |
Giant Golden Orbweaver spider |
Nephila pilipes |
Araneidae |
OQ780848 |
631 bp |
Least concern |
|
35 |
Cape Rainspider |
Palystes castaneus |
Sparassidae |
OQ821217 |
648 bp |
Not evaluated |
|
36 |
Joro spider |
Trichonephila clavata |
Araneidae |
OQ821219 |
658 bp |
Least concern |
|
37 |
Kogane-gumo spider |
Argiope amoena |
Araneidae |
OQ821220 |
658 bp |
Not evaluated |
|
38 |
Brown widow spider |
Latrodectus geometricus |
Theridiidae |
OQ780838 |
617 bp |
Not evaluated |
|
39 |
Ant |
Platythyrea punctata |
Formicidae |
OQ780845 |
619 bp |
Not evaluated |
|
40 |
Ant |
Messor ebeninus |
Formicidae |
OQ780846 |
612bp |
Not evaluated |
|
41 |
Common sailor butterfly |
Neptis hylas |
Nymphalidae |
OQ821228 |
658 bp |
Not evaluated |
|
42 |
Gypsy moth |
Lymantria dispar |
Erebidae |
OQ821229 |
654 bp |
Not evaluated |
|
43 |
Small ant |
Tapinoma sessile |
Formicidae |
OQ780847 |
624 bp |
Not evaluated |
|
44 |
Common batwing butterfly |
Atrophaneura varuna |
Papilionidae |
OQ821232 |
658 bp |
Least concern |
|
45 |
Tropical swallowtail moth |
Lyssa zampa |
Uraniidae |
OQ825991 |
658 bp |
Not evaluated |
|
46 |
Giant peacock moth |
Saturnia pyri |
Saturniidae |
OQ825992 |
658 bp |
Not evaluated |
|
47 |
Brown tussock moth |
Olene mendosa |
Erebidae |
OQ825993 |
658 bp |
Not evaluated |
|
48 |
Thief ant |
Solenopsis molesta |
Formicidae |
OQ825994 |
576 bp |
Not evaluated |
|
49 |
Northern warrior wasp |
Synoeca septentrionalis |
Vespidae |
OQ780849 |
626 bp |
Least concern |
|
50 |
Giant Asian mantis |
Hierodula patellifera |
Mantidae |
OQ825995 |
658 bp |
Not evaluated |
|
51 |
Common hairy caterpillar |
Spilarctia obliqua |
Erebidae |
OQ825989 |
658 bp |
Least concern |
|
52 |
Kaempfer cicada |
Platypleura kaempferi |
Cicadidae |
OQ825986 |
685 bp |
Not evaluated |
|
53 |
Giant shield bugs |
Pycanum ochraceum |
Tessaratomidae |
OQ825985 |
630 bp |
Not evaluated |
|
54 |
Shield bugs |
Pentatoma metallifera |
Pentatomidae |
OQ825983 |
641 bp |
Not evaluated |
|
55 |
Leopard lacewing butterfly |
Cethosia cyane |
Nymphalidae |
OQ825990 |
642 bp |
Not evaluated |
|
56 |
Black soldier fly |
Hermetia illucens |
Stratiomyidae |
OQ825987 |
658 bp |
Not evaluated |
|
57 |
Silver- spotted skipper butterfly |
Epargyreus clarus |
Hesperiidae |
OQ825988 |
658 bp |
Least concern |
|
58 |
Cicada |
Okanagana rimosa |
Cicadidae |
OQ780833 |
624 bp |
Not evaluated |
|
59 |
Grasshopper |
Dociostaurus maroccanus |
Acrididae |
OQ780836 |
590 bp |
Not evaluated |
|
60 |
Tobacco grasshopper |
Atractomorpha crenulata |
Pyrgomorphidae |
OQ780837 |
597 bp |
Not evaluated |
|
61 |
Southern green stink bug |
Nezara viridula |
Pentatomidae |
OQ825984 |
658 bp |
Least concern |
|
62 |
Florida woods cockroach |
Eurycotis floridana |
Blattidae |
OQ780841 |
630 bp |
Not evaluated |
|
*Consevation status |
||||||
The species identified included, 20 birds, 11 mammals, 5 spiders, 2 lizards and 24 insects. From the present studied locations, Tragopan blythii (Jerdon, 1870), Sitta formosa (Blyth, 1843) and Rusa unicolor (Kerr, 1792) were reported and declared as Vulnerable species by IUCN.3.1 (Red List, 1964). These species are declining due to overexploitation, traditional Jhum shifting cultivation and hunting activities.
 |
Figure 2: Insects species from study areas identified using molecular markers.
|
 |
Figure 3: Spiders species from study areas identified using molecular markers.
|
 |
Figure 4: Lizards and birds species from study areas identified using molecular markers.
|
 |
Figure 5: Mammals species from study areas identified using molecular markers.
|
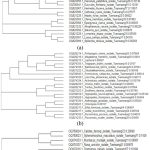 |
Figure 6: The evolutionary relationships of: a, insects and spiders; b, birds; and c, mammals and lizards taken for molecular study.
|
All the DNA barcoding sequences were aligned and used to create phylogenetic trees informing us of the evolutionary relationships among closely related species, as shown in Figure 5. Extensive research has shown that DNA barcoding can identify a wide range of animal species such as mammals, reptiles, birds, fishes, amphibians and crustaceans12,28,30. One of the most effective and advanced methods for identifying insects is DNA barcoding, which uses the mitochondrial gene cytochrome c oxidase I (COI) sequence as a DNA identifier10. The sequences discovered will help to clarify the evolutionary history, for example, of spider mites and will enable quick species identification using molecular methods11. DNA barcodes cannot replace the need for taxonomists, but DNA barcodes provide an additional suite of characters that can be used in taxonomy, assisting identification of species by non-specialists with accurate identification of their specimens. When it comes to saving animals under difficult conditions, genetic information is extremely valuable13.
A newly detailed understanding of species diversity can illuminate processes important in speciation, as suggested by the discovery that the most diverse lineages of animals. By using various molecular markers, it is possible to detect unique genetic variation in endangered population or species4. Knowledge of genetic diversity directs us to develop breeding programs minimizing inbreeding and safeguarding the loss of genetic variation. It is evident that the combination of morphological and molecular data sets permits a clearer recognition of evolutionary diversity among organisms. Further study is necessary to reveal the diversity among family members14. Applications of the mitochondrial gene-based species identification techniques cover a wide spectrum, and offer a fascinating perspective for future animal study6,15.
The mitochondrial gene cytochrome c oxidase I (COI) barcode reference library for animals has become a standard resource for DNA metabarcoding applications recommending as standard metabarcode for metazoans3. These advances have opened up numerous new applications in aquatic sciences and ecological monitoring27. Still, further work is needed to optimize use of COI barcodes for these applications5.
Conclusion
Molecular markers are very useful, providing information on genetic variability among and within animal populations, helping us to develop appropriate conservation strategies. Choosing an efficient profiling technique is important, as selection of an inappropriate technique may lead to incorrect conservation actions. It is important to highlight, however, that DNA barcoding should not replace traditional taxonomy, as integrating both methods can provide a more thorough knowledge of species diversity. The current study improved our knowledge of the faunal diversity of selected areas of Tuensang district, Nagaland. Non-taxonomists, researchers, biodiversity managers and policymakers will use the information provided to strengthen their efforts to develop efficient protective measures for this faunal biodiversity.
Acknowledgment
The author would like to thank the Head, Department of Zoology, St.Joseph University, Dimapur, Nagaland for her guidance and support.
Conflict of Interest
The authors declare no conflict of interest.
Funding Source
There are no funding sources.
Authors’ Contribution
Chaueichongla Phom (Research scholar) and Dr. Jeyaparvarthi Somasundaram (Supervisor)
Data Availability Statement
Not applicable
Ethics Approval Statement
The study does not involves an experiment on humans and animals. No Ethical approval was conducted.
References
- Abobakr Y., Al-Sarar A.S, Alzabib A.A and Saleh A.A. Morphological and molecular characterization of the invasive pestiferous land snail Macrochlamys indica Godwin-Austen, 1883 (Gastropoda: Ariophantidae) from Saudi Arabia. Agriculture. 2022; 12(11): 1756.
CrossRef - Abubakar B.M., Salleh F. M, Omar M S and Wagiran A. DNA barcoding and chromatography fingerprints for the authentication of botanicals in herbal medicinal products. Evidence-Based Complementary and Alternative Medicine. 2017; 2017:1352948.
CrossRef - Andújar C., Arribas P., Yu D.W.,Vogler A.P and Emerson B.C. Why the COI barcode should be the community DNA metabarcode for the metazoa. Ecol. 2018; 27: 3968–3975.
CrossRef - Asmelash B., Diriba S and Pal S. K. Molecular markers based characterization and conservation of wild animals. Research Journal of Recent Sciences. 2017; 6(7):53-62.
- Collins R.A., Bakker J., Wangensteen O.S., Soto A.Z., Corrigan L., Sims D.W., Genner M.J., Mariani S. Non-specific amplification compromises environmental DNA metabarcoding with COI. Methods Ecol. Evol. 2019; 10:1985–2001.
CrossRef - Elyasigorji Z., Izadpanah M., Hadi F and Zare M. Mitochondrial genes as strong molecular markers for species identification. Oxidative Stress in Applied Basic Researh and Clinical Practice, 2023; 66:81-93.
CrossRef - Hebert P.D.N., Cywinska A., Ball S.L and deWaard J.R. Biological identifications through DNA barcodes. Proc Biol Sci. 2003; 270(1512): 313–321.
CrossRef - Hebert P.D.N., Ratnasingha S., Zakharov E.V., Telfer A., Levesque-Beaudin V., Milton M.A., Pedersen S., Jannetta P., deWaard J.R. Counting animal species with DNA barcodes: Canadian insects. Philosophical Transactions of the Royal Society B: Bilolgical Sciences. 2015; 371(1702): 0150333.
CrossRef - Hendrich L, Moriniere J, Haszprunar G, Hebert PDN, Hausmann A, Kohler F, Balke M. A comprehensive DNA barcode database for Central European beetles with a focus on Germany: adding more than 3500 identified species in BOLD. Mol. Ecol. Res. 2015; 15: 795–818.
CrossRef - Hussain, K., Rashid, K., Hafeez, F, Amad I and Ashraf M.Molecular identification of sugarcane black bug (Cavelarius excavates) from Pakistan using cytochrome C oxidase I (COI) gene as DNA barcode. Int J Trop Insect Sci., 2020; 40: 1119–1124.
CrossRef - İnak E., Çobanoğlu S., Auger P and Migeon A.Molecular identification and phylogenetic analysis of spider mites (Prostigmata: Tetranychidae) of Turkey. Exp Appl Acarol., 2022; 87:195–205.
CrossRef - Ivanova N.V., Clare E.L and Borisenko A.V. DNA barcoding in mammals. Methods in Molecular Biology. 2012; 858: 153–182.
CrossRef - Jayasundara S L, Algewatta H R, Jayawardana S, Perera M and Peiris L D C. Molecular identification and evolutionary divergence of the Sri Lankan sambar deer, Rusa unicolor (Kerr 1792). Animals, 2023; 13: 2877.
CrossRef - Johansen J.R., Bohunická M., Lukešová A., Hrčková K., Vaccarino M.A and Chesarino N.M. Morpgological and molecular characterization within 26 strains of the Genus Cylindrospermum (Nostocaceae, Cynobacteria),with descriptions of three new species. Phycol., 2014; 50(1):187-202.
CrossRef - Malewski T, Dzikowski A and Soltyszewski I. Molecular methods of animal species identification. Polish Journal of Natural Science, 2021; 36(1):79-94.
- Mani M.,Venkatesan T., and Chethan B.R. Molecular identifications of insects pests of horticultural crops. Trends in Horticulture Entomology, 2022; 978-981-19-0343-4.
- Mayr E. Speciation and macroevolution. International Journal of Organic Evolution. 1982; 36(6):1119-1132.
CrossRef - Mishra P., Kumar A., Nagireddy A., Mani D. N., Shukla A. K.,Tiwari R and Sundaresan V. DNA barcoding: an efficient tool to overcome authentication challenges in the herbal market. Plant Biotechnology Journal. 2015; 14(1): 8–21.
CrossRef - Mushtaq H.M., Selah A.A., Kamran M and Alatawi F.J. Molecular-based taxonomic inferences of some spider mite species of the genus Oligonchus Berlese (Acari, Prostigmata, Tetranychidae. Insects. 2023;14:192.
CrossRef - Ratnasingham S and Hebert P.D.N. A DNA-based registry for all animal species: the Barcode Index Number (BIN) system. PLoS ONE. 2013; 8(7): e66213.
CrossRef - Sara M. F, Cristina S.L. , Moreira I and Felah B.S.A.B. DNA barcoding of commercially relevant marine fish species in Tunisian waters. Journal of the Marine Biological Association of the United Kingdom. 2022; 102(3-4): 178 – 185.
CrossRef - Schmidt S., Schmidt-Egger C., Moriniere J., Haszprunar G., Hebert P.D.N. DNA barcoding largely supports 250 years of classical taxonomy: Identifications for Central European bees (Hymenoptera: Apoideapartim). Ecol. Res. 2015; l5: 985–1000.
CrossRef - Shashank P.R., Naveena N.L., Rajgopal N.N., Elliott T.A, Sreedevi K, Sunil S and Meshram N M. DNA barcoding of insects from India: Current status and future perspectives. Mol Biol Rep., 2022; 49:10617–
CrossRef - Smith T.B and Wayne R.K. Molecular Genetic Approaches in Conservation. Oxford University Press, New York. 1996; 101:504.
CrossRef - Szaro R.C and Johnston D.W. Biodiversity in mangrove landscapes, theory and practice. Biodiversity Conservation. 1996; 72(1):778 .
- Tzudir L and Markandan M. Identification and documentation of edible insects of Vespa affinis in Nagaland by using DNA barcode techniques. International Journal of Pharmaceutical Sciences and Research, 2021; 12(3): 1529-1532.
- Valentini A., Taberlet P., Miaud C., Civade , Herder J., Thomsen P. F., Bellemain E., Besnard A., Coissac E., Boyer F., et al. Next-generation monitoring of aquatic biodiversity using environmental DNA metabarcoding. Mol. Ecol. 2016; 25:929–942.
CrossRef - Vences M., Nagy Z. T., Sonet G and Verheyen E. DNA barcoding amphibians and reptiles. Methods in Molecular Biology. 2012; 858: 79–107.
CrossRef - Wan Q.H., Wu H., Fang S.G and Fujihara T. Which genetic marker for which conservation genetics issue? Electrophoresis. 2004;25(14):2165-2176.
CrossRef - Yang F., Ding F., Chen H., He M., Zhu S., Ma X., Jiang L and Li H. DNA Barcoding for the identification and authentication of animal species in traditional medicine. Evidence-Based Complementary and Alternative Medicine. 2018; 5160254: 18.
CrossRef

This work is licensed under a Creative Commons Attribution 4.0 International License.





