Manuscript accepted on : 27-11-2023
Published online on: 12-12-2023
Plagiarism Check: Yes
Reviewed by: Dr. Nidhi Trivedi
Second Review by: Dr Maysaloun Abdulhameed AL-Sadoon
Final Approval by: Dr. Wagih Ghannam
Dasari Thrimothi * , Edla Sujatha
, Edla Sujatha , Kuraganti Guna Swetha and Gudikandula Krishna
, Kuraganti Guna Swetha and Gudikandula Krishna
Department of Microbiology, Kakatiya University, Warangal, Telangana, India.
Corresponding Author E-mail: trimothidasari2286@gmail.com
DOI : http://dx.doi.org/10.13005/bbra/3177
ABSTRACT: One of the extracellular enzymes involved in ligninolytic is lacasse, secreted by white and brown rot fungus. The class of blue copper proteins that includes laccases consists of N-glycosylated multicopper oxidases. Ascomycetes, Deuteromycetes, and Basidiomycetes are all fungi that contain laccase; many of these are white-rot fungi that break down lignin. Due to their wide range of substrate specificity, laccases have been the focus of extensive research over the past few decades. Their latest applications include anything from the textile pulp and paper industries to culinary applications and bioremediation techniques. Laccases are also used in organic synthesis, where phenols and amines are common substrates. Dimers and oligomers are produced due to the coupling of reactive radical intermediates in these reactions. The current investigation gathered 50 white rot fungi, and the most incredible laccase-producing organisms in submerged fermentation were looked into. Ten of the 21 cultures displayed a reddish-brown color zone. Of these ten isolates (PTD 19, PTD 4, PP2J15, LKT 34, ITC 1, NRL 7, GOJ 7, PTD2, PP2J, and PKT12), only PP2J15 and GOJ 7 displayed the most reddish-brown color zone. The isolation of white rot fungus, their molecular characterization, and testing for laccase production are all covered in this Paper. Talaromyces verruculosus and Cladosporium cladosporioides were identified as the PP2J15 and GOJ 7 strains based on sequence comparison and phylogenetic analysis with reference taxa.
KEYWORDS: Guaiacol, Laccase; PDA; Telangana; Wood rot fungi
Download this article as:| Copy the following to cite this article: Thrimothi D, Sujatha E, Swetha K. G, Krishna G. Isolation, Screening, Identification, and Assessment of Laccase-Producing Fungi Isolated From Different Environmental Samples. Biotech Res Asia 2023;20(4). |
| Copy the following to cite this URL: Thrimothi D, Sujatha E, Swetha K. G, Krishna G. Isolation, Screening, Identification, and Assessment of Laccase-Producing Fungi Isolated From Different Environmental Samples. Biotech Res Asia 2023;20(4). Available from: https://bit.ly/3RFO6wr |
Introduction
A class of fungi known as the Basidiomycetes is renowned for producing a variety of extracellular ligninolytic enzymes. The benzenediol: oxygen oxidoreductases (EC 1.10.3.2) laccases (Lac) and different peroxidases, such as lignin peroxidase (LiP), manganese-dependent peroxidase (MnP), and versatile peroxidase (VP), are among these enzymes. While MnP is classed as a Mn (II): hydrogen peroxide oxidoreductase (EC 1.11.1.13), LiP is a diarylpropane: oxygen, hydrogen peroxide oxidoreductase (C-C-bond-cleaving) enzyme 1-3. VP demonstrates both LiP and MnP catalytic capabilities. The enzymes under investigation are essential for the lignin breakdown in their native lignocellulosic substrates and the xenobiotic dye breakdown 4. While some fungi that break down wood have all three types of lignin-altering enzymes, others only have one or two 5. White-rot fungi produce lignin-modifying enzymes as part of their secondary metabolism 6. The enzymes discussed in this context are frequently created and secreted in response to carbon or nitrogen constraints 7. High oxygen tension is often the best environment for MnP and LiP enzyme synthesis. However, agitation in liquid cultures of submerged white-rot fungi inhibits this process. However, agitation, aromatic chemicals, or organic solvents are routinely used to increase the production of Lac 8. In aerated cultures, without adding oxygen and subjected to shaking conditions, the enzyme VP, previously identified as a manganese-oxidizing enzyme capable of oxidizing veratryl alcohol, is produced 9. They were investigating white-rot fungi and their enzymes to see whether they could be used to break down various aromatic contaminants that cause environmental problems. Specifically, the pollutants examined included those found in pulp and paper mills [10], olive mill wastewater 11, polycyclic aromatic hydrocarbons 10, chlorinated phenols, polychlorinated biphenyls 12, dioxins, pesticides, explosives, and dyes 13. It is typical for numerous isoforms of ligninolytic enzymes to be expressed in distinct taxa and culture conditions. The characteristics above are crucial for the process design and fungal treatment of effluent optimization. For various commercial applications, purified Lac, LiP, and MnP enzymes show significant potential 14. The extraordinary capacity of fungi to synthesize different extracellular enzymes is well known. Basidiomycetes are the primary organisms that break down lignocellulose 15. White-rot, brown-rot, soft-rot, and litter-decomposing fungi are three frequent groups of basidiomycetous fungi that break down wood 16, 17. The only organisms that can successfully mineralize lignin are related to litter-decomposing fungi and basidiomycetous white-rot fungi 5.
Recently, there has been a growing interest in researching the lignin-modifying enzymes in various white-rot fungi. This interest derives from a comparative biological viewpoint and the hope of finding lignin-degrading systems that are more effective and can be applied in multiple biotechnological applications. The availability of information on the production of extracellular oxidoreductases by native fungal strains from various eco-physiological and taxonomic categories is, however, limited.
The three goals of this study were to (i) Collection of fungi from different Tunisian biotopes representing a variety of eco-physiological and taxonomic groups, (ii) To assess the ligninolytic potential of the fungal strains, (iii) To improve the cultivation conditions that favor high laccase yield from white-rot fungi.
Material and Methods
Isolation and maintenance of Pure Culture
To isolate fungus, fifty samples of soil were dug up from each site at a depth of 5 to 10 cm, and 100 g of the sample was taken using a sterile spatula and placed in pre-sterilized zip-lock bags. In Telangana, ecological forest soil and industrial effluents were the sources of the samples (Table 1 and Fig. 1). Pure cultures of isolated fungal species were produced using a potato dextrose agar (PDA) medium. Using the dilution plate method, one gram of each soil sample was extracted and put into a test tube with 10 ml of sterile distilled water [18]. The suspension was serially diluted in sterile distilled water five times, from 10-1 to 10-5. After vigorously mixing the soil samples, the solution was obtained. After letting the suspension stand for 20 minutes, 0.5ml of each aqueous dilution of the soil suspension was inoculated on Petri plates containing Potato dextrose agar. To prevent bacterial contamination, the PDA medium was given a trace quantity of streptomycin. Techniques like spread plate and pour plate were employed. For six days, the inoculation plates were incubated at 30 °C. To create pure cultures, several fungal colonies were sub-cultured (Table 1).
A small amount of fungus spores (or) fruit body was transferred from an active fungus culture to brand-new test tubes (screw cap or clogged with cotton or foam) or Petri dishes (covered with Parafilm to reduce drying) containing an appropriate agar medium. A culture is maintained at room temperature once it has been created. Periodically inspect cultures for contamination. Every three weeks, these are subcultured, tested for purity, and then subcultured again. Slants and plates were used to preserve pure cultures. There were 21 pure cultures recovered from the 50 samples collected throughout different regions in Telangana. The laccase enzyme was estimated using these pure cultures. The diverse collection of organisms known as white-rot fungi can break down lignin, other types of wood, contaminated soil, and phenolic chemicals. The non-specific and non-stereo selective extracellular enzyme systems are responsible for the capacity to break down lignin. Laccases make for the extracellular enzyme system that breaks down lignin. Because the essential elements of the whit-rot lignin-degrading system are extracellular, the fungi can break down dangerous environmental contaminants and insoluble compounds like lignin.
Table 1: Sample collection sites
| Site No | Site Name | Strain Code | Source | Altitude |
| 1 | Perfect Tanners, Warangal | PTD | Leather processed effluent | 17.9990° N, 79.6242° E |
| 2 | ITC, Bhadrachalam, Sarapaka | ITC | Paper and pulp-processed effluent | 17.6943° N, 80.8628° E |
| 3 | Pakala Wildlife Sanctuary Forest | PKT | Leaf litter, decaying wood | 17° 56′ 59.99″ N 79° 58′ 59.99″ E |
| 4 | Laknavaram Forest | LKT | Digged soil sample, decaying wood | 18°3′ 52.0092” N 79°29’39.4548” E |
| 5 | Mythri Rubber Industries, Rampoor | MRT | Excretory effluent | 17.94541° N, 79.45780° E |
| 6 | Autonagar, Warangal | ATG | Dumped soil | 18.00536° N, 79.59800° E |
| 7 | Nagaram Lake | NLT | Polluted water sample | 18.08856° N, 79.57042° E |
| 8 | Pembathi-contaminated paddy soil, | PP2J | Contaminated soil | 18.08493° N, 79.53934° E |
| 9 | Nerella forest soil | GOJ | Digged soil sample | 18.15037° N, 79.57170° E |
 |
Figure 1: Sample collection from various Environmental sites. |
Results
Qualitative assay for Laccase enzyme
To choose appropriate generating strains, screening laccase enzyme-producing white rot fungus species was crucial. Due to the many characteristics involved, screening for oxidative enzymes or mediators necessitates the examination of numerous samples. This is why the current method employs low-cost, quick, and sensitive testing techniques. The screening technique must be designed to find fungal strains and usually functioning enzymes. Using a guaiacol plate assay, the 21 pure cultures were examined for the efficiency of their laccase activity. Guaiacol (0.02%) was added to the Potato Dextrose Agar medium, which was then incubated for six days at 30°C with pure colonies. This laccase enzyme estimate is qualitative. Guaiacol contained in the media was able to be oxidized by white rot fungus that generated the laccase enzyme, producing a reddish-brown zone (Fig. 2). Fig. 3 demonstrated that the absence of a reddish-brown color zone in the medium suggests that the laccase enzyme synthesis is not taking place. The zone was measured and shown in Table 2 for comparison. It was clear from the table that ten fungal cultures out of the 21 pure cultures displayed a reddish-brown color zone on medium. This suggests that guaiacol is utilized as a marker for extracellular oxidative enzymes, which is supported by the current finding. Ten of them (PTD 19, PTD 4, PP2J15, LKT 34, ITC 1, NRL 7, GOJ 7, PTD2, PP2J, and PKT12) had the reddish-brown color zone from the start. The red-brown zone in the white rot fungal cultures was highest in PP2J15 at 7 mm at three days and 10 mm at six days, whereas GOJ7 displayed reddish-brown zone measurements of 6 mm at three days and 9 mm at six days.
Nayak et al. (2022) adopted a similar strategy and employed the guaiacol plate assay method for qualitative screening. Fungal isolates were point inoculated onto the corresponding potato dextrose agar plate that contained guaiacol (0.02%) and were then incubated at 28°C for six days. The families Chaetomiaceae, Pleosporaceae, Trichocomaceae, Nectriaceae, and Ajellomycetaeae were identified by morphological research on isolated fungi 19. The qualitative study on Potato Dextrose Agar plates with 0.02% guaiacol was reported by Anitha et al. in 2022. The plates were incubated at 30°C for five days. After five days of incubation, the laccase-producing fungal strains displayed a reddish-brown halo around the colonies on the Guaiacol-containing plates 20. Nayak et al. (2017) qualitatively evaluated twenty-three lignocellulolytic fungi using a guaiacol plate assay on potato dextrose agar medium. Five fungal isolates out of 23 fungal strains produced the most quality laccase 21. A similar experiment was carried out by Gnanasalomi and Gnanadoss (2013) to identify the laccase-producing fungus. It was carried out on solid media containing colored markers such as guaiacol, 2,2 -azinobis (3-ethylbenzthiazoline-6-sulphonic acid), syringaldazine, and polymeric dyes 22. According to Desai et al. (2011), the growth of laccase-producing fungi on potato dextrose agar (PDA) media containing tannic acid and guaiacol PDA plates were observed for growth and development of reddish-brown colored precipitate in plates and observed reddish hallow zone in guaiacol PDA plates 23.
The screening is based on the indicators’ color changes, which are connected to the activity of the ligninolytic enzymes. Tanwar et al. (2020) qualitative screening for visualizing corresponding hydrolysis zones of several isolates, revealing their capacity to generate diverse carbohydrates, including laccase, cellulase, and xylanase, was published in 2020. Six of the 31 isolates with cellulase activity had the largest zone size, whereas six of the 31 with xylanase activity had the largest zone size. Multiple hydrolytic enzyme activities of laccase, cellulase, and xylanase were present in ligninolytic fungal isolates, and different strains were chosen based on the zones of hydrolysis around the fungal colonies that were the highest 24.
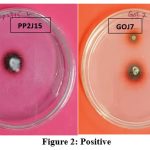 |
Figure 2: Positive Click here to view Figure |
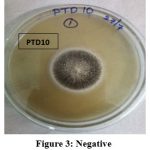 |
Figure 3: Negative Click here to view Figure |
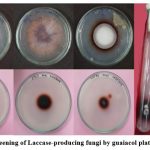 |
Figure 4: Screening of Laccase-producing fungi by guaiacol plate technique. Click here to view Figure |
 |
Figure 5: In the medium, the reddish-brown color zone has not formed. Click here to view Figure |
Table 2: Qualitative assay of laccase enzyme by white rot fungi.
| S. No | Fungi | Conc. of Guaiacol
(µl) |
3rd DAY
Diameter of reddish-brown zone (mm) |
6th DAY
Diameter of reddish-brown zone (mm) |
| 1 | PTD 19 | 20µl | 3mm | 5mm |
| 2 | PTD 4 | 20µl | 4mm | 6mm |
| 3 | PP2J15 | 20µl | 7mm | 10mm |
| 4 | LKT 34 | 20µl | 3mm | 5mm |
| 5 | ITC 1 | 20µl | 5mm | 6mm |
| 6 | CM10 | 20µl | — | — |
| 7 | NRL7 | 20µl | 1mm | 3mm |
| 8 | PM5J | 20µl | 2mm | 4mm |
| 9 | CM1 | 20µl | 3mm | 4mm |
| 10 | PTD5 | 20µl | — | — |
| 11 | GOJ7 | 20µl | 6mm | 9mm |
| 12 | PTD2 | 20µl | 2mm | 3mm |
| 13 | LKT11 | 20µl | 1mm | 2mm |
| 14 | LKT10 | 20µl | 4mm | 6mm |
| 15 | PTD6 | 20µl | — | — |
| 16 | CM3 | 20µl | 2mm | 5mm |
| 17 | PP2J | 20µl | 3mm | 7mm |
| 18 | MRT2 | 20µl | — | — |
| 19 | ITC12 | 20µl | 2mm | 3mm |
| 20 | PKT12 | 20µl | 3mm | 6mm |
| 21 | ATG5 | 20µl | 4mm | 6mm |
Quantitative assay for laccase enzyme
To estimate laccase enzyme levels, the fungal isolates were cultivated in potato dextrose broth as pure cultures. For up to 12 days, pure cultures were cultured in potato dextrose broth. For the sixth and twelfth days of incubation, laccase enzyme activity was determined using a UV-visible spectrophotometer, and pH changes were noted using a pH meter. Ten of the 21 fungi examined for laccase activity in fungal cultures revealed positive results, while the other 14 were negative.
The two cultures with the highest output of laccase enzyme were PP2J15 and GOJ7 (Table 3). The laccase activity in the fungi isolates PP2J15 was notable, reaching 625 U/ml on day 6th and 712 U/ml on day 12th. GOJ7 displayed laccase activity of 273 U/ml on day 6th and 512 U/ml on day 12th. For additional activities, these two cultures were utilized. On plates with guaiacol supplementation and the development of colored zones during laccase activity screening, a strong link between the two was discovered.
Pranitha et al. (2015) employed a similar strategy. Pure fungal isolates were collected and cultured in malt extract broth to determine enzyme estimates. Malt extract broth was incubated for up to 14 days after being inoculated with pure colonies. 18 of the 30 fungi tested for lignolytic activity out of 30 exhibited positive results, and the rest tested negative. Pv5 demonstrated, among the cultures, the most significant production of lignolytic enzymes. Pv5 showed laccase activity 350 U/ml on 7th day, 606 U/ml on 14th day, LiP activity 412 U/ml on 7th day, 544 U/ml on 14th day, MnP activity 126 U/ml on 7th day and 102 U/ml on 14th day 25. In another study, Tapwal et al. (2014) reported four fungal isolates (ANF36, ANF212, ANF218, and ANF238) produced brown coloration around their colony and were positive for guaiacol plate assay 26. Selvam et al. (2012) employed a novel method to confirm the lignolytic activity of the fungus. Guaiacol is one of the most often used substrates for qualitative and quantitative laccase assessment. The capacity to grow on and degrade lignin was investigated in a lignin-amended basal medium. The growth was quantified as an increase in mycelia dry weight (mg/day). The % of lignin degradation was determined to be in the range of 20.4 to 68.0, and the mycelial growth rate was in the field of 1.24 mg/day to 3.67 mg/day 27.
Table 3: Quantitative assay of laccase enzyme by collected white rot fungi
| S. No | Fungi | pH | Laccase U/ml | ||
| 6th | 12th | 6th | 12th | ||
| 1 | PTD 19 | 4 | 4.3 | 211 | 36 |
| 2 | PTD 4 | 5 | 3.6 | 66 | 121 |
| 3 | PP2J15 | 5 | 5.5 | 625 | 712 |
| 4 | LKT 34 | 4.5 | 4.5 | 76 | 56 |
| 5 | ITC 1 | 5 | 5 | 152 | 182 |
| 6 | CM10 | 3 | 4 | — | — |
| 7 | NRL7 | 5 | 4.5 | 136 | 98 |
| 8 | PM5J | 3 | 5 | 12 | 2 |
| 9 | CM1 | 5 | 5.8 | 22 | 46 |
| 10 | PTD5 | 3.5 | 4.1 | – | – |
| 11 | GOJ7 | 4 | 5.1 | 273 | 512 |
| 12 | PTD2 | 5 | 4.5 | 62 | 74 |
| 13 | LKT11 | 4 | 4 | 144 | – |
| 14 | LKT10 | 5 | 5.5 | – | – |
| 15 | PTD6 | 4 | 3.2 | 38 | 67 |
| 16 | CM3 | 5 | 4.5 | – | – |
| 17 | PP2J | 3 | 3.5 | 124 | 4 |
| 18 | MRT2 | 5 | 4 | 46 | 98 |
| 19 | ITC12 | 5 | 4 | 36 | 48 |
| 20 | PKT12 | 5 | 5.5 | 197 | 149 |
| 21 | ATG5 | 5 | 5 | – | – |
Selection of Organism
Two white rot fungi demonstrated the highest levels of lignolytic activity according to the results of quantitative and qualitative tests, i.e., Talaromyces verruculosus (OR782472.1) and Cladosporium cladosporioides (OR649270), which were selected for further studies.
Molecular Identification
Talaromyces verruculosus (OR782472.1)
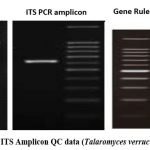 |
Figure 6: gDNA and ITS Amplicon QC data (Talaromyces verruculosus (OR782472.1). Click here to view Figure |
Sequences producing significant alignments
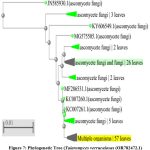 |
Figure 7: Phylogenetic Tree (Talaromyces verruculosus (OR782472.1) Click here to view Figure |
Table 4: Table representing the identified species with their respective accession number while sequence alignment with culture (OR782472.1).
| Accession | Description |
| MG551571.1 | Talaromyces verruculosus isolate 567 |
| KC506174.1 | Fungal sp. AM2013 strain 7 |
| KY263515.1 | Talaromyces verruculosus strain PPRI 14967 |
| HQ657296.1 | Penicillium sp. CRCF6 18S ribosomal RNA gene |
| KJ767054.1 | Talaromyces siamensis isolate A3S2-39 |
| MW344647.1 | Talaromyces verruculosus isolate RK4Tm |
| OQ076506.1 | Penicillium sp. isolate CF00120 |
| KJ482652.1 | Talaromyces verruculosus strain AG67 |
| OW988114.1 | Talaromyces siamensis genomic DNA sequence |
| OP295495.1 | Talaromyces australis strain RR1331 |
Talaromyces verruculosus (OR782472.1) is a member of Ascomycota with the following classification.
Domain: Eukaryota
Kingdom: Fungi
Phylum: Ascomycota
Class: Eurotiomycetes
Subclass: Eurotiomycetidae
Order: Eurotiales
Family: Trichocomaceae
Genus: Talaromyces
Species: T.verruculosus
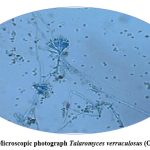 |
Figure 8: Microscopic photograph Talaromyces verruculosus (OR782472.1). Click here to view Figure |
Cladosporium cladosporioides (OR649270)
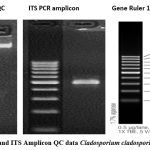 |
Figure 9: gDNA and ITS Amplicon QC data Cladosporium cladosporioides (OR649270). Click here to view Figure |
 |
Figure 10: Phylogenetic Tree Cladosporium cladosporioides (OR649270). Click here to view Figure |
Sequences producing significant alignments:
Table 5: Table representing the identified species with their respective accession number while sequence alignment with culture (OR649270)
| Accession | Description |
| OR244405.1 | Cladosporium cladosporioides |
| OR135243.1 | Cladosporium cladosporioides isolate 10395-2022 |
| OQ443091.1 | Cladosporium cladosporioides isolate DHY8 |
| MN429197.1 | Cladosporium tenuissimum isolate F8144 |
| FJ613828.1 | Fungal endophyte sp. ZY-2009 |
| MK367508.1 | Cladosporium sp. isolate Z-Y-20 |
| KU508795.1 | Cladosporium cladosporioides strain CSPF3 |
| KP689176.1 | Cladosporium cladosporioides isolate FL20 |
| ON248258.1 | Cladosporium sp. isolate QTY4 |
| KF907245.1 | Dothideomycetes sp. UOM H |
Cladosporium cladosporioides (OR649270) is a member of Ascomycota with the following classification.
Domain: Eukaryota
Kingdom: Fungi
Phylum: Ascomycota
Class: Dothideomycetes
Subclass: Dothideomycetidae
Order: Capnodiales
Family: Davidiellaceae
Genus: Cladosporium
Species: C.cladosporioides
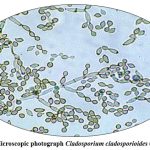 |
Figure 11: Microscopic photograph Cladosporium cladosporioides (OR649270). Click here to view Figure |
Discussion
Fungi have the unique capability to efficiently break down wood components among all microbes. In the present study, various fungal strains capable of generating laccase were recovered from diverse environmental samples. The primary goals of the current study were to identify and classify laccase-producing fungi from Telangana’s (Forest soil sample, industrial excretory) forest litter and industrial effluents. Twenty-one fungi were found in industrial effluents and forest trash. The preservation of microbial isolates was accomplished using Potato Dextrose Agar. To identify laccase-producing fungus derived from fungi isolated from diverse environmental samples, a straightforward screening technique was employed. This method involved the utilization of solid media containing the indicator component guaiacol. Guaiacol was used as a substrate for qualitative screening of their potential laccase manufacturing ability in the future. All of the fungal isolates were added to the potato dextrose agar plates containing 0.02% guaiacol and then cultured for five days at 30°C. The results of screening tests are displayed in Table 2. After 3 to 6 days of incubation, the fungal isolates acquired a reddish-brown halo surrounding the colonies, which indicates that laccase was produced; the absence of a reddish-brown halo indicates that laccase was not produced.
The Guaiacol plate test revealed good laccase activity in the two elite fungal isolates Talaromyces verruculosus (OR782472.1) (7mm on the third day, 10mm on the sixth day) and Cladosporium cladosporioides (OR649270) (6mm at third day, 9mm at sixth day). Incubation of the Talaromyces verruculosus (OR782472.1) and Cladosporium cladosporioides (OR649270) elite fungal isolates in Potato dextrose medium at room temperature resulted in maximal laccase production of 625 U/ml at day six, 712 U/ml at day twelve, and 273 U/ml at day six, 512 U/ml at day twelve, respectively. The molecular characterization of top fungal isolates is shown in Figures 7 and 10. Similar research was conducted by Singh et al. (2013), who also screened laccase producers like Trichoderma harzianum on potato dextrose agar supplemented with 0.04% guaiacol. They isolated laccase-producing fungi on PDA plates containing 0.02% of guaiacol and observed a reddish-brown oxidation zone [28]. According to Senthivelan et al. (2019), using guaiacol as a substrate, a UV-visible spectrophotometer was used to assess the laccase activity of the white rot fungus, which was found to be 3.2 U/ml [29]. According to Monssef et al. (2016), Trichoderma harzianum produced laccase at a rate of 1.479 U/ml as determined by a UV-visible spectrophotometer with guaiacol as the substrate [30].
Conclusion
To choose strains that can manufacture laccase enzyme in the current investigation, a screening of several ambient and soil samples taken from the forest ecosystem is required. Ten isolated cultures out of the twenty-one distinct cultures were determined to be laccase-positive. In order to identify an influential laccase producer from the separate cultures, quantitative estimation was used. The isolate PP2J15 was discovered to be the most effective laccase producer. Talaromyces verruculosus was eventually determined to be the strain (OR782472.1) based on morphological traits and 18S rRNA gene sequencing. Maximum laccase production was recorded at days 6 and 12, respectively, and isolate (OR649270) was later determined to be Cladosporium cladosporioides based on morphological traits and 18S rRNA gene sequencing—maximum laccase production (273 U/ml on day 6 and 512 U/ml on day 12). A strain’s potential for commercial laccase production would not solely depend on its capacity to produce the laccase enzyme. In order to increase the productivity of the target enzyme for a range of biotechnological applications, parametric optimization must be carried out to determine optimal growth and production conditions for the chosen strain. The current study also indicates the capacity of a unique ecological group of Ascomycota-phylum litter-dwelling/decomposing fungi (LDF) to produce lignin-modifying enzymes, including laccase and Mn-peroxidase. LDF was determined to be the better degrader of lignin and lignin-related chemicals despite lower titers of ligninolytic enzymes. LDF is a possible substitute for traditional white rot fungi because they do not produce enough ligninolytic enzymes or show sufficient growth in a competitive setting like soil litter.
Acknowledgement
The authors are grateful to the Head, Department of Microbiology, Kakatiya University, Warangal for the continuous encouragement and support to carry out this research work. The authors also thank the technical staff for their valuable services to perform the work.
Conflict of Interest
The authors declare that there is no conflict of interests regarding the publication of this paper.
Funding Sources
There is no funding source
Reference
- Rabha J, Devi SP, Das S, Roy N, Jha DK. Microbial conversion of biomass to value-added chemicals. Value-Addition in Agri-food Industry Waste Through Enzyme Technology. 2023 Jan 1:37-64.
CrossRef - Pozdnyakova NN, Balandina SA, Dubrovskaya EV, Golubev CN, Turkovskaya OV. Ligninolytic basidiomycetes as promising organisms for the mycoremediation of PAH-contaminated Environments. InIOP Conference Series: Earth and Environmental Science 2018 Jan 1 (Vol. 107, p. 012071). IOP Publishing.
CrossRef - Hatti‐Kaul R, Ibrahim V. Lignin‐Degrading Enzymes: An Overview. Bioprocessing technologies in biorefinery for sustainable production of fuels, chemicals, and polymers. 2013 Jul 12:167-92.
CrossRef - Singh D, Gupta N. Microbial laccase: a robust enzyme and its industrial applications. Biologia. 2020 Aug;75:1183-93..
CrossRef - Mir Tutusaus JA. Process development for hospital wastewater treatment by Trametes versicolor.
- Heri SM. Functional characterization of lignin modifying enzymes produced by white rot fungi for their mycoremediation potential of petroleum (xenobiotic) polluted soil(Doctoral dissertation) 2022.
- Gmoser R, Ferreira JA, Lennartsson PR, Taherzadeh MJ. Filamentous ascomycetes fungi as a source of natural pigments. Fungal biology and biotechnology. 2017 Dec;4:1-25.
CrossRef - Kuhad RC, Kuhar S, Sharma KK, Shrivastava B. Microorganisms and enzymes involved in lignin degradation vis-à-vis production of nutritionally rich animal feed: an overview. Biotechnology for environmental management and resource recovery. 2013:3-44.
CrossRef - Husain Q, Husain M. Peroxidases as a potential tool for the decolorization and removal of synthetic dyes from polluted water. Environmental protection strategies for sustainable development. 2012:453-98.
CrossRef - Rodríguez-Couto S. Industrial and environmental applications of white-rot fungi. Mycosphere. 2017 Mar;8(3):456-66.
CrossRef - Adenipekun CO, Lawal R. Uses of mushrooms in bioremediation: A review. Biotechnology and Molecular Biology Reviews. 2012 Sep 30;7(3):62-8.
CrossRef - Jurado M, Martinèz ÀT, Martinez MJ, Saparrat MC. 6.45 Application of white-rot fungi in transformation, detoxification, or revalorization of agriculture wastes: role of laccase in the processes. Comprehensive Biotechnology. 2011;6:595-603.
CrossRef - Zhang J, Chi Y, Feng L. The mechanism of degradation of alizarin red by a white-rot fungus Trametes gibbosa. BMC biotechnology. 2021 Nov 5;21(1):64.
CrossRef - Mtui GY. Lignocellulolytic enzymes from tropical fungi: Types, substrates and applications. Res. Essays. 2012 Apr 23;7(15):1544-55.
CrossRef - Sharma HK, Xu C, Qin W. Biological pretreatment of lignocellulosic biomass for biofuels and bioproducts: an overview. Waste and Biomass Valorization. 2019 Feb 15;10:235-51.
CrossRef - Eichlerová I, Baldrian P. Ligninolytic enzyme production and decolorization capacity of synthetic dyes by saprotrophic white rot, brown rot, and litter decomposing Basidiomycetes. Journal of Fungi. 2020 Nov 19;6(4):301.
CrossRef - Mali T, Mäki M, Hellen H, Heinonsalo J, Bäck J, Lundell T. Decomposition of spruce wood and release of volatile organic compounds depend on decay type, fungal interactions and enzyme production patterns. FEMS Microbiology Ecology. 2019 Sep;95(9):fiz135.
CrossRef - Shinde RE, Shahi DK, Mahapatra P, Naik SK, Singh CS, Verma SH, Singh AK. Isolation of lignocelluloses degrading microbes from soil and their screening based on qualitative analysis and enzymatic assays. Plant Soil Res. 2022;24:347-54.
CrossRef - Nayak B, Choudhary R, MG R. Isolation, Screening and Morphological characterization of Laccase producing fungi. International Research Journal on Advanced Science Hub. 2022 Feb 28;4(2):38-43.
CrossRef - Anitha K, Asha K, Prasad G, Bayineni VK, Rahaman AH, et al. Isolation and Screening of Laccase Producing Fungi from Different Regions of Western Ghats. J Agric Forest Meteorol Res. 2022, 5(2): 422-427.
- NAYAK B, CHOUDHARY R, ROYMOM M. LIGNOCELLULOLYTIC FUNGAL ISOLATION AND SCREENING FOR THEIR LACCASE PRODUCING ABILITY. Indian J. Sci. Res. 2017;13(2):188-91.
- Gnanasalomi VD, Gnanadoss JJ. Laccase production by Myrothecium gramineum and its optimization under solid state fermentation using cowpea pod as substrate. Journal of Applied Biology and Biotechnology. 2019 Mar 5;7(2):59-63.
CrossRef - Desai SS, Tennali GB, Channur N, Anup AC, Deshpande G, Murtuza BA. Isolation of laccase producing fungi and partial characterization of laccase. Bioinf. Bioeng. 2011;1(4):543-9.
- Tanwar D, Sharma N, Sharma N, Prusty PK. Isolation and screening of fungi from rotten wood for various hydrolytic enzymes production. Annals of Phytomedicine. 2020;9(1):122-8.
CrossRef - Vadapally P, Gudikandula K, Maringanti SC. Isolation, Screening and Identification of Laccase Producing Fungi from Eturnagaram Forest, Warangal District, Telangana, India. Science, Technology and Arts Research Journal. 2015;4(1):120-3.
CrossRef - Tapwal A, Varghese S, Kumar U, Arora J. Production of Laccase by Alternaria alternata and Lasiodiplodia theobromae. European Journal of Experimental Biology. 2014;4(4):196-201.
- Selvam K, Shanmuga Priya M, Sivaraj C, Arungandhi K. Identification and Screening of wood rot fungi from Western Ghats area of South India. Int J Chem Tech Res. 2012;4:379-88.
- Singh MP, Vishwakarma SK, Srivastava AK. Bioremediation of direct blue 14 and extracellular ligninolytic enzyme production by white rot fungi: Pleurotus spp. BioMed Research International. 2013 Jan 1;2013.
CrossRef - Senthivelan T, Kanagaraj J, Panda RC, Narayani T. Screening and production of a potential extracellular fungal laccase from Penicillium chrysogenum: Media optimization by response surface methodology (RSM) and central composite rotatable design (CCRD). Biotechnology Reports. 2019 Sep 1;23:e00344.
CrossRef - Abd El Monssef RA, Hassan EA, Ramadan EM. Production of laccase enzyme for their potential application to decolorize fungal pigments on aging paper and parchment. Annals of Agricultural Sciences. 2016 Jun 1;61(1):145-54.
CrossRef

This work is licensed under a Creative Commons Attribution 4.0 International License.





