Manuscript accepted on : 25-05-2023
Published online on: 20-06-2023
Plagiarism Check: Yes
Reviewed by: Dr. Farah Ramzi Nouri
Second Review by: Dr. Robert Susło
Final Approval by: Dr. Eugene A. Silow
Molecular approaches in soil microbial analysis: Forensic Perspective
Preeti Sangwan1 , Tarsem Nain2
, Tarsem Nain2 , Priyanka Yadav1
, Priyanka Yadav1 and Neelkamal Sharma1*
and Neelkamal Sharma1*
1Department of Forensic Science, Maharshi Dayanand University, Rohtak, Haryana India.
2Department of Genetics, Maharshi Dayanand University, Rohtak, Haryana India.
Corresponding Author E-mail: neelforensics@gmail.com
DOI : http://dx.doi.org/10.13005/bbra/3096
ABSTRACT: The growing field of soil microbial forensics provides the legal answer to how microorganisms play a crucial role in criminal investigations. It is an advanced cross-disciplinary science capable of offering significant physical evidence with considerable interest in criminal investigations, environmental crime, and counterterrorism. Microbial forensics of soil consists of different techniques to identify and evaluate microbial abundance, their complexity, and their interaction with soil and surrounding objects. The present review highlights various microbial analysis techniques such as Terminal restriction fragment length polymorphism, Temperature/Denaturing Gradient Gel Electrophoresis, Amplified Ribosomal DNA Restriction Analysis, Length Heterogeneity PCR, Phospholipid-derived fatty acids, Fluorescence in situ hybridization, Stable-isotope probing and metagenomics using next-generation sequencing. This article also summarises the challenges faced in soil microbial forensics, various statistical approaches, reference databases commonly used in forensic soil studies, and different methodological approaches used in forensic laboratories. Literature was studied using various online bibliographic databases like Google Scholar, Web of Science, Pub Med, Scopus, and several other search engines. Conclusive evidence generated by this paper signifies the importance of soil microbes to assist forensic scientists and researchers in selecting adequate methods to differentiate soil samples. The present comparative study concluded that targeted molecular analysis of microbes as a forensic soil typing tool has a lot of potential and should be investigated further.
KEYWORDS: DNA; Forensic; Metagenomics; Microbes; Soil
Download this article as:| Copy the following to cite this article: Sangwan P, Nain T, Yadav P, Sharma N. Molecular approaches in soil microbial analysis: Forensic Perspective. Biosci Biotech Res Asia 2023;20(2). |
| Copy the following to cite this URL: Sangwan P, Nain T, Yadav P, Sharma N. Molecular approaches in soil microbial analysis: Forensic Perspective. Biosci Biotech Res Asia 2023;20(2). Available from: https://bit.ly/46fWGHA |
Introduction
Forensic Pedology is the science that uses soil information to solve officially approved questions, problems, or hypotheses 1. Soil is more likely to transfer and retained as it is typically found on clothes, footwear, and vehicles following a crime. It could be a potent method of contact as physical evidence, especially when criminals tend to forget soil evidence while tempering other pieces of evidence 2.
During the early 1990s, soil or sediment as physical evidence gained great attention 3-5. The soil and sediment analysis was used in forensic investigations by Prof. Ehrenberg in the 19th century to solve the missing silver coin mystery and effectively settle crime 2. In 1887, Sir Arthur Connan Doyle published many fictional novels involving soil comparison to solve the murder by Sherlock Holmes. In 1904, George Popp, a forensic scientist, skilfully studied soil, mineral contents, dirt, and debris from clothing to determine criminal cases 6. The soil analysis is also effectively utilised as evidence in legal proceedings in Australia, the United Kingdom, and the USA 7.
Soil forensics is an emerging discipline that needs the standardization of novel methods to construct a database so that soil can be utilised as physical evidence in criminal investigations. Forensic scientists frequently employ a systematic methodology for screening purposes based on soil colour, soil texture, consistency, particle size, pH, and density 8. After this, spectroscopic techniques were used for the elemental analysis of the soil 9-12. After the initial screening and spectroscopic analysis of soil, more robust and statistically reliable methods are required for microbial profiling. This research presents a more detailed picture of microbial methods, including DNA fingerprinting methods and metagenomics, to identify a specific microbial community in the soil.
Soil microbial forensics is the science that can define how microbial communities can be used in criminal enquiries 13. Microbes are clearly a substantial soil constituent, with one tablespoon of soil having roughly 109 microorganisms. Merely 1% of soil microbes can be cultured by traditional methods, which makes soil one of the most complex ecosystems requiring molecular techniques to assess them 14.
Soil microbial communities
The (micro) bio-composition of soil significantly enlightens the new field of soil microbial forensics 15. Microbes can convey information about the specific ecological environment that sustains them. The authors claimed that soil might be distinct due to the presence of specialised microbes present in the soil 16.
Each community may be present at a given density based on the soil type. Physical, chemical, biological, environmental, and anthropogenic factors can significantly impact the variety and diversity of soil microorganisms, which are unique for the particular soil type to be sampled 17.
Materials and methodology
In this review, attempts have been made to document the importance of soil microbes to assist forensic scientists and researchers in selecting adequate methods to differentiate soil samples. Recent literature was cited by conducting a thorough search of electronic databases, such as PubMed, ScienceDirect, Web of Science, Scopus, and Google Scholar, by using appropriate/ specific combinations of words.
Molecular approach for the soil’s microbial diversity
The two primary categories of approaches used to explore soil microbial diversity are molecular and biochemical methods. However, current molecular tools are gaining more attention for crime investigations than biochemical methods due to their precision, sensitivity, feasibility and early results 18. In the present review, we emphasised the utilisation of molecular technologies for forensic investigations.
DNA-based analysis of soil using DNA fingerprinting techniques
The forensic community uses several analysis methods to generate soil DNA profiling, but none are specifically designed for forensic use. Therefore, the forensic community must pick an approach that best meets its specific needs. DNA fingerprinting techniques that evaluate fragment length variation comprise Terminal Restriction Fragment Length Polymorphism (T-RFLP) 19, 20, Denaturing Gradient Gel Electrophoresis (DGGE) 21, 22 and Temperature Gradient Gel Electrophoresis (TGGE) analysis, Amplified Ribosomal DNA Restriction Analysis (ARDRA) 23 and Length Heterogeneity- Polymerase Chain Reaction (LH-PCR) 18, 24.
Terminal Restriction Fragment Length Polymorphism (T-RFLP)
The T-RFLP approach, first introduced by Liu et al. 25 in 1997, has been acknowledged as a quick and effective way to create or monitor modified changes in the “structure and composition” of microbial communities 26. This approach is based on various fragment length that provides a unique pattern (fingerprint) liable bio-composition of the species present in the sample. T-RFLP fingerprinting method could be used to generate DNA profiles from the small amount of soil used to differentiate the samples 19.
Macdonald et al. 27 explored the application of multiplex T-RFLP as a tool for soil microbes’ comparison from a forensic point of view. This technique employed the combined benefits of multiple taxa with the T-RFLP approach, which offers a fast and cost-effective study of a microbial population at high-resolution. Macdonald et al. 28 investigated the ability of T-RFLP for microorganisms to discriminate between soils of different sites and exhibited clear differentiation, which may be helpful for location identification. Various forensic investigation of soil microbiota profiling has employed T-RFLP because it has high reproducibility and automatic nature 19, 29, 30
T-RFLP is a valuable tool for the basic evaluation of soil microbes’ population. Resolution and taxonomic identification are limited by the co-migration of many taxa during electrophoresis, resulting in displays as a single band 31. Because of this, additional analysis through complex methods like next-generation sequencing, which has enough discriminatory power to identify soil microbes, is required for the sample that seems to possess the same T-RFLP profile but may not necessarily originate from a common source 32. Another drawback of this approach is library dependency, which necessitates the design of a library database for bacteria, archaea, and fungi.
Denaturing gradient gel electrophoresis (DGGE) and temperature gradient gel electrophoresis (TGGE) analysis
DGGE and TGGE are gel-based methods, employing either chemical or temperature gradients that denature DNA samples while passing across an acrylamide gel. This method can be utilised with DNA and RNA samples and proteins but is less widely used. DGGE distinguishes genes of the same size based on their various denaturing potentials, which are defined by the arrangement of their base pairs.
A study published by Lerner et al. 21 described the use of DGGE to conduct criminal investigations and has enough discriminating potential to discriminate between variations in soil microbiota’s temporal and spatial variability. In one example, a young woman’s corpse was found near the river bank. After some days, the primary suspect was apprehended and claimed to be with the victim in the parking lot. After committing the crime, the suspect washed his clothes and shoes. There was no proof except a small amount of soil inside the suspect’s shoes. DGGE method was conducted on the samples taken from the crime scene, alibi scene, the accused’s house, and the surrounding areas. With the help of cluster analysis, banding characteristics were assessed. The samples gathered from the crime scene, and its environs were grouped, whereas those from the alibi scene and its surrounding could easily be differentiated from the crime scene. Furthermore, Sanachai et al.33 elucidated that the origin of soil, obtained from the sole of a shoe, could be identified by a similarity comparison of soil bacterial 16S rDNA profiles separated by the DGGE method.
Amplified Ribosomal DNA Restriction Analysis (ARDRA)
This fingerprinting technique is a robust tool for bacterium identification 34 and for investigating bacterial diversity in soil microorganisms 35. Horswell et al. 36 investigated a case of forensic soil identification in which they employed ARDRA to analyse 16S rDNA of soil bacteria by detecting the fluorescent labelled terminal fragment of RFLP (T-RFLP). In an investigation into a murder case in northern Italy, Concheri et al. 23 used the ARDRA method to compare or match the similarity of soil samples taken from the crime scene and the carpet of a suspect’s car. The results showed that the soil found in the car matched the soil taken from the crime scene. The results of this research had a significant role in the court’s decision. The ARDRA method was also used by Naknim et al. 37 to compare soil evidence from shoes collected from a mock crime scene with soil from the mock crime scene as well as irrelevant areas, and results showed that ARDRA is a reliable method for identifying the origin of soil by comparing soil bacterial community structure by clustering of 16S rDNA restriction profiles.
Length Heterogeneity- Polymerase Chain Reaction (LH-PCR)
Length Heterogeneity PCR (LH-PCR), a modified version of the PCR method, is used extensively in several microbiology fields and is gaining prominence in the field of soil microbial analysis. LH- PCR differentiates different microorganisms based on naturally occurring sequence length of DNA 24, 38. Moreno et al. 18 compared the microbial metagenome profiles generated using LH-PCR analysis of 16S rRNA genes with Inductive Coupled Plasma- Optical Emission Spectroscopy (ICP-OES) analysis of 13 elements commonly found in soils. The findings revealed that microbial metagenome profiling was better than chemical characterisation could discriminate between various soil types and had a high reproducibility, proving a potential tool for soil comparisons in the criminal investigation.
This fingerprinting method helps predict geographical locations and provides an investigative tool that the suspect spent time at a specific site. A brilliant example of a study design for the prediction of location is provided by Damaso et al. 39. The Authors studied soil DNA profiles using LH-PCR method to test the biogeographical patterns of soils to determine whether soil microbial community is spatially correlated with a geographic location or not. Moreover, the results found that soil microbial communities have unique patterns and are spatially auto-correlated.
Phospholipid Fatty Acids in Soils (PLFAs)
PLFAs are the key component present in the soil microbe cell membrane. PLF analysis is the strategic biotechnological tool that shows significant differences in cellular membrane compositions of 2 different microbial communities for forensic soil discrimination application purposes 40. PLFA analysis collected from the various soil sample provides efficient structural information on the microbial community. PLFA also provide information about the pattern of fatty acids present and the total microbial biomass of the soil microbial community 41. It is relatively inexpensive, reproducible, highly precise, lowest error rate and rapid methods employ certain advantages viz. community-level physiological profiling 42 over DNA-based (DNA fingerprinting, Electrophoresis) methods 43.
Fluorescence in situ hybridization (FISH)
Individual soil microbial cells can be simultaneously recognized, counted, localized and discriminate by using Fluorescence In Situ Hybridization (FISH) advanced analytical technique 44. FISH is a non-fingerprinting and highly sensitive technique due to the specificity to work on the low amount of rRNA 45’ 46. It is a frequently used, strong and effective technique in differentiating the microbial profiles of several soil samples with related geological properties 47.
Stable Isotope Probing (SIP)
SIP is a biomedical technique used for the tracking of the movement of nutrients from isotopically tagged substrates to particular microbes in microbial communities. Stable-Isotope Probing, or SIP, continues to be one of the most comprehensive methods for in situ microbial community analysis 48. To reconstruct the metagenome-assembled genomes (MAGs) of the microorganisms that produce tagged proteins, proteomic SIP and targeted metagenomic binning were used 49. Using proteomic SIP, active complex microbial communities from different soil samples collected from different places are functionally characterised in forensic prospectives.
Cases studies of soil microbial evidence
Around the world, several cases have been solved utilising soil evidence by using microbial DNA analysis techniques (Table 1).
Table 1: Some case studies solved using soil microbial community profiling.
| Types of cases | Evidence | Country | Method of analysis | References
|
| Murder case | Suspect’s shoes | Israel | DGGE | 21
|
| Murder case | Car carpet and tyres | Italy | ICP-MS, ICP-OES and ARDRA | 23 |
| Mock case study | Shoes, shovel, car tyres | Australia | HTS and MIR spectroscopy | 50 |
| Mock crime scene | shoes | Thailand | ARDRA | 37 |
| Looting of a burial vault | A pair of green and brown boots | Spain | Colour, particle size distribution, elemental analysis, anion concentration, pH, rDNA 16S sequencing
|
51 |
| Rape case | Shoes, shovel | Not specified | tRFLP profiling and pollen analysis | 7
|
| Mock crime scene (drug burial case) | Spade | Not specified | RISA and 16S rRNA gene sequencing with Illumina MiSeq | 52 |
| Missing and murder case | Socks | Arizona | Illumina MiSeq | 53 |
| Simulated crime scene | Shoe and sampling tool | China | HTS of 16S rRNA gene | 54 |
Metagenomics
The term metagenomics was invented in 1998 by Handelsman et al. Metagenomics (also called environmental genomics, eco-genomics, or community genomics) is the study of genetic material derived from a diverse community of organisms that are used to provide taxonomic and functional profiles of soil microbes 55. It is a molecular tool used to analyse DNA obtained from soil samples to study the soil microbes’ community without obtaining pure culture. Metagenomics allows an understanding of the different characteristics of a sample, characterises microbes, and describes the functional roles of environmental soil microbes present in the samples. The use of metagenomics has steadily increased and provided new forensic identification opportunities 56, 57.
The primary aim of these techniques is to rebuild large-scale genomic data or functional processes of a selection of their genes. Several newer culture-independent metagenomics approaches do not use DNA, such as amplicon-based, whole metagenome-based, and functional-based metagenomic analysis.
16S and 18S rRNA are the most common genes sequenced and amplified in the soil microbe’s community. Both genes offer valuable information about the diversity and abundance of bacteria and archaea (16S rRNA) and eukaryotes as fungi (18S rRNA). Above mentioned sequences are the most conserved and represent microbial genetic dissimilarity. Therefore, it indicates fundamental differences in phyla, genera, and species. This is the primary tool for identifying and characterising the soil microbial community, which may be valuable for soil microbial forensics in criminal investigations 58. It is a reliable and cost-effective way to differentiate microbial communities from thousands of samples.
Metagenomics using Next Generation Sequencing (NGS)
NGS is a high-throughput sequencing method used for metagenomics analysis of the diverse microbial population, including their metabolic potential, structure, and effects on ecosystem function. Before NGS technology, forensic experts could not work with microbes because sequencing methods were too slow and expensive or dependent on culture-based methods. But with the advent of NGS technology, experts can identify DNA sequences of every microorganism present in a sample accurately, rapidly and comprehensively 59, and avoid experimental contamination caused by microbial cultures, which has proven helpful in forensics 60.
The use of NGS tools has increased in such studies over the past decade as technologies have evolved from the 454 Roche and MiSeq Illumina to Nanopore and SMRT PacBio. 61 compared non-culturing dependent tools and concluded that NGS can be combined with PLFA (PhosphoLipid Fatty-acid Analysis) to get a structural and functional picture of the entire microbial community in soil.
Numerous studies have been carried out by microbiologists to explore soil microbes using a next-generation sequence, but there is little research from a forensic point of view. Researchers surveyed soil microbes’ diversity and solved a fictional case study using a next-generation sequencing-based on 16s and 18sRNA genes, plant chloroplast leucine tRNA gene and fungal spacer region between rRNA genes 62, 63. The authors evaluated the effect of the development of fungal profiles through next-generation sequencing. The findings suggested that the development of the fungal profile was unaffected by the quantity of soil. Even tiny traces of soil, typically encountered in forensic case studies, provided valid genetic details 64. The researchers distinguished very similar and dissimilar habitat types over time and space and soil on evidence items by assessing bacterial 16s rRNA gene through next-generation sequencing. Forensic studies have also examined statistical approaches to accurately evaluate the large sequencing datasets to classify and differentiate soil samples 65, 66. Finley et al. 67 provided comprehensive literature about potential soil microbes ecology and NGS applications for forensic purposes.
Advantages and disadvantages of molecular-based methods
Each molecular method has its advantages and disadvantages. An overview is given in Table 2.
Table 2: Advantages and disadvantages of various molecular-based methods used in studying soil microbial community.
| Approach | Molecular methods | Advantages | Disadvantages | References |
| DNA Fingerprint techniques | T-RFLP | · High sensitivity.
· Effective at analysing the relationship of bacterial communities in diverse ecological samples · The capability of high throughput and microbial community quantification · Short run time. |
· Overestimation of diversity due to incomplete restriction digestion.
· Complex profiles make phylogenetic assignments very difficult. · Required a diverse range of restriction enzymes to explore microbial diversity. |
68-70 |
| DGGE/TGGE | · For amplification and sequencing, bands of interest can be excised from the gel. | · Limited sensitivity.
· Handling gels need experience. · DNA sequences of different bacterial species can exhibit the same separation as a result of equivalent GC contents. |
71, 72 | |
| ARDRA | · Fast, accurate and straightforward molecular tool to determine environmental population profile.
· No special equipment is needed. |
· Low discriminatory power as compared to other fingerprinting methods.
· Needs multiple restrictions for adequate genotypic resolution. · Difficulty in locating a particular phylogenetic group within a community fingerprint. |
73, 74 | |
| LH-PCR | · Monitor bacterial populations in diverse settings.
· Fast and allow simultaneous analyses of multiple and complex samples. |
· Shows the high number of secondary peaks. | 75 | |
| DNA sequencing techniques | 454 Pyrosequencing | · Estimation of soil microbial diversity at a vast geographical scale.
· The maximum number of unique sequences can be identified. |
· Highly conserved primers are used to amplify hypervariable regions.
· Expensive equipment. · High background signal after every cycle. · Difficulties in the sequencing of GC-rich templates. |
76, 77 |
| Illumina (Mi Seq model) | · Simple, scalable and high yield.
· Have a much faster turnaround time. |
· Low throughput molecular techniques.
· Expensive equipment. |
78 | |
| Ion Torrent | · Generate and read sequences from both ends of a fragment.
· Low cost and fast run. |
· High rate of sequencing errors. | 77 |
Statistical analysis tools for multivariate data
Although various methods for routine investigations have been developed and characterised, the findings must be evaluated with suitable and reliable statistical methods. Molecular techniques such as T-RFLP, DGGE, FAME, FISH, DNA sequencing, etc., provide complex data interpreted through specific statistical methods. The statistical tools viz., SIMPER 79, ANOSIM 80, PCA 81-83, and Cluster analysis 84 are used to identify and individualise the component from multivariate data.
Reference databases for soil microbial community analysis
Soils have high microbial diversity and variability, which challenges studying microbial communities. To improve the understanding of the soil microbial community, we require a reference database or tools. Various tools and software are used for the soil microbial analysis for evaluation and interpretation such as Greengenes 85, SILVA 86, 87, RDP (Ribosomal Database Project) 88, NCBI (National Center for Biotechnology Information) 89, MG-RAST (Metagenomic Rapid Annotations using Subsystems Technology) 90, 91, IMG/M (Integrated Microbial Genomes and Metagenomes) 92, CAMERA 93, GOLD (Genomes OnLine Database) 94, MEGAN (Metagenomic Analyzer) 95 and RefSoil 96.
Sequential examination of soil evidence in forensic laboratories
Soils are extremely diverse, complex, and external disturbance and pollution can alter physicochemical and microbial content; therefore, it is necessary to manage, store and transport to the forensic laboratory properly. The microbial analysis of soil in forensic laboratories has five steps: (1) collection, storage, and transportation of soil samples from the crime scene; (2) analysis by using the molecular tool, metagenomic sequencing, and classification of taxa; (3) comparison of the findings with the database; (4) evaluation and interpretation of the results and last (5) presenting the findings to the court (Fig. 1).
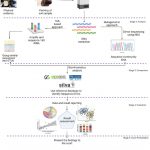 |
Figure 1: Diagrammatic representation of microbial analysis of soil evidence from the crime scene to the court. |
(Although the author made the figure, the inspiration for it came from the source ref. 67).
Challenges faced in soil microbial forensics
There are several challenges faced in soil microbial forensics, some of which include:
Sample collection and preservation
Soil samples need to be collected and preserved correctly to prevent contamination or degradation of the microbial community. Improper handling of the soil samples can lead to false results, making it challenging to determine the source of the microorganisms.
Microbial diversity
Soil contains a vase and a diverse microbial community that can vary depending on the location, time of year, and other factors. Therefore, it can be challenging to identify and differentiate microbial populations, making it difficult to trace the source of the microorganisms 97.
Limited reference databases
There is limited availability of microbial reference databases that can be used to compare and identify microbial populations. This makes it difficult to match soil samples to a specific location or source 98.
Environmental factors
Soil microbial populations can be affected by various environmental factors, such as temperature, moisture, and pH levels. These factors can cause variations in the microbial community and make it challenging to determine the origin of the microorganisms 99.
Legal challenges
Soil microbial forensics is a relatively new field, and there is limited legal precedence for the use of microbial evidence in court cases. Therefore, there may be challenges in presenting microbial evidence in a court of law.
Overall, soil microbial forensics is a complex field that requires specialised knowledge and techniques to overcome the challenges. Advances in technology and collaboration among experts can help address some of these challenges and improve the accuracy and reliability of soil microbial forensics.
Conclusive findings and discussion
Soil microbial forensics, an emerging field that exploits the diversity and dynamics of soil microorganisms, has great potential for various applications, such as environmental monitoring, agriculture, and forensics100. Through application of advanced techniques, the article summarizes the novel strategies related to the composition, spatial patterns, and ecological roles of soil microbiomes in forensic prospectives. Diverse group of techniques focused on identifying microbial signatures that can be used to differentiate between soils from different geographic regions or environments101. These signatures may include specific microbial taxa, functional genes, or metabolic pathways.
Nowadays significant advancements in the methods used to study soil microbial communities, including the development of high-throughput sequencing techniques and bioinformatics tools for analysing microbial data99. Despite the potential of soil microbial forensics, some significant challenges and limitations need to be addressed. These include issues related to reproducibility and standardization of methods, as well as the need for a better understanding of the factors that influence microbial community composition in soil. Cited literature highlights the development of new methods for analysing soil microbial communities, as well as further investigations into the potential applications of soil microbial forensics in different fields. The review summarizes the current state-of-the-art molecular methods for soil microbial profiling and characterization and discusses the challenges and future directions of this field.
Conclusion
The diverse, complex, and heterogeneous nature of soil makes it a reliable resource for linking culprit-victim relations in the criminal field. Forensic science and legal courts’ proceedings are progressively relying on the study of DNA for crime resolution. Soil offers a wealth of DNA data that needs to be implemented to the full extent and value. The use of molecular methods for forensic purposes is novel and holds excellent potential by offering forensic scientists an additional ‘tool in the toolbox’. This molecular approach proves to be a powerful and differentiating toolbox for the molecular exploration of different soil microbial samples with identical geographical features. The purpose of this study is to explore the geopolitical location of the crime site, help in intelligence work, locate clandestine burials, estimate Post-mortem Interval (PMI), reduce the search area and comparative analysis of microbial community that supports the evidence when used in court.
Acknowledgement
The authors would like to thank University Grants Commission (UGC), New Delhi for providing financial assistance. Grant number: 3422/(NET-NOV 2017)
Conflict of Interest
Authors declare no conflict of interest.
Funding Sources
This study was funded by University Grant Commission, New Delhi (3422/(NET- NOV 2017)).
References
- Stella TWL, Swarup S, See Suet Ning M, Lim NQBI, Phua Mun Lin S, Tan Boon Jay T, et al. Forensic Pedology: From Soil Trace Evidence to Courtroom. Soil Analysis: Recent Trends and Applications: Springer; 2020. p. 305-23.
- Fitzpatrick RW. Soil: forensic analysis. Wiley encyclopedia of forensic science2009. p. 1-14.
- Hiraoka Y. A possible approach to soil discrimination using X-ray fluorescence analysis. Journal of Forensic Science. 1994;39(6):1381-92.
- Junger E. Assessing the unique characteristics of close-proximity soil samples: just how useful is soil evidence? Journal of Forensic Science. 1996;41(1):27-34.
- Lee B, Williamson T, Graham R, Lund L. Forensic soils: an integrative laboratory exercise for introductory soil science. Journal of Natural Resources and Life Sciences Education. 1998;27(1):110-2.
- Ruffell A, McKinley J. Forensic geoscience: applications of geology, geomorphology and geophysics to criminal investigations. Earth-Science Reviews. 2005;69(3-4):235-47.
- Uitdehaag S, Quaak F, Kuiper I. Soil comparisons using small soil traces, a case report. Soil in Criminal and Environmental Forensics: Springer; 2016. p. 61-9.
- Marumo Y. Forensic examination of soil evidence. Japanese Journal of Science and Technology for Identification. 2003;7(2):95-111.
- Cox R, Peterson H, Young J, Cusik C, Espinoza E. The forensic analysis of soil organic by FTIR. Forensic Science International. 2000;108(2):107-16.
- Pye K, Croft D. Forensic analysis of soil and sediment traces by scanning electron microscopy and energy-dispersive X-ray analysis: An experimental investigation. Forensic Science International. 2007;165(1):52-63.
- Xu X, Du C, Ma F, Shen Y, Zhou J. Forensic soil analysis using laser-induced breakdown spectroscopy (LIBS) and Fourier transform infrared total attenuated reflectance spectroscopy (FTIR-ATR): principles and case studies. Forensic Science International. 2020;310:110222.
- Li T, Song F, Zhang J, Liu S, Xing B, Bai Y. Pyrolysis characteristics of soil humic substances using TG-FTIR-MS combined with kinetic models. Science of the Total Environment. 2020;698:134237.
- Santiago-Rodriguez TM, Cano RJ. Soil microbial forensics. Microbiology Spectrum. 2016;4(4):4.. 30.
- Zala K. Forensic science. Dirty science: soil forensics digs into new techniques. Science (New York, NY). 2007;318(5849):386-7.
- Ruffell A. Forensic pedology, forensic geology, forensic geoscience, geoforensics and soil forensics. Forensic Science International. 2010;202(1-3):9-12.
- Efeoğlu FG, Çakan H, Kara U, Daş T. Forensic Microbiological Analysis of Soil and the Physical Evidence Buried in Soil Obtained from Several Towns in Istanbul. Cureus. 2022;14(2):e22329.
- Yang T, Lupwayi N, Marc S-A, Siddique KH, Bainard LD. Anthropogenic drivers of soil microbial communities and impacts on soil biological functions in agroecosystems. Global Ecology and Conservation. 2021;27:e01521.
- Moreno LI, Mills DK, Entry J, Sautter RT, Mathee K. Microbial metagenome profiling using amplicon length heterogeneity‐polymerase chain reaction proves more effective than elemental analysis in discriminating soil specimens. Journal of Forensic Sciences. 2006;51(6):1315-22.
- Gryta A, Frąc M. Methodological aspects of multiplex terminal restriction fragment length polymorphism-technique to describe the genetic diversity of soil bacteria, archaea and fungi. Sensors. 2020;20(11):3292.
- Gałązka A, Grządziel J. The molecular-based methods used for studying bacterial diversity in soils contaminated with PAHs (The Review). Soil Contamination-Current Consequences and Further Solutions. 2016:85-104.
- Lerner A, Shor Y, Vinokurov A, Okon Y, Jurkevitch E. Can denaturing gradient gel electrophoresis (DGGE) analysis of amplified 16s rDNA of soil bacterial populations be used in forensic investigations? Soil Biology and Biochemistry. 2006;38(6):1188-92.
- Pasternak Z, Al-Ashhab A, Gatica J, Gafni R, Avraham S. Optimization of molecular methods and statistical procedures for forensic fingerprinting of microbial soil communities. Int Res J Microbiol. 2012;3(11):363-72.
- Concheri G, Bertoldi D, Polone E, Otto S, Larcher R, Squartini A. Chemical elemental distribution and soil DNA fingerprints provide the critical evidence in murder case investigation. PLoS One. 2011;6(6):e20222.
- Mills DK, Entry JA, Gillevet PM. Assessing microbial community diversity using amplicon length heterogeneity polymerase chain reaction. Soil Science Society of America Journal. 2007;71(2):572-8.
- Liu W-T, Marsh TL, Cheng H, Forney LJ. Characterization of microbial diversity by determining terminal restriction fragment length polymorphisms of genes encoding 16S rRNA. Applied and Environmental Microbiology. 1997;63(11):4516-22.
- Sabale SN, Suryawanshi PP, Krishnaraj P. Soil metagenomics: concepts and applications. Metagenomics-Basics, Methods and Applications: IntechOpen; 2019.
- Macdonald LM, Singh BK, Thomas N, Brewer MJ, Campbell CD, Dawson LA. Microbial DNA profiling by multiplex terminal restriction fragment length polymorphism for forensic comparison of soil and the influence of sample condition. Journal of Applied Microbiology. 2008;105(3):813-21.
- Macdonald CA, Ang R, Cordiner SJ, Horswell J. Discrimination of soils at regional and local levels using bacterial and fungal T‐RFLP profiling. Journal of Forensic Sciences. 2011;56(1):61-9.
- Habtom H, Demanèche S, Dawson L, Azulay C, Matan O, Robe P, et al. Soil characterisation by bacterial community analysis for forensic applications: a quantitative comparison of environmental technologies. Forensic Science International: Genetics. 2017;26:21-9.
- Kandasamy S, Liu EYR, Patterson G, Saldias S, Ali S, Lazarovits G. Introducing key microbes from high productive soil transforms native soil microbial community of low productive soil. Microbiologyopen. 2019;8(10):e895.
- Young J, Austin J, Weyrich L. Soil DNA metabarcoding and high-throughput sequencing as a forensic tool: considerations, potential limitations and recommendations. FEMS Microbiology Ecology. 2017;93(2).
- Thies JE. Soil microbial community analysis using terminal restriction fragment length polymorphisms. Soil Science Society of America Journal. 2007;71(2):579-91.
- Sanachai A, Katekeaw S, Lomthaisong K. Forensic soil investigation from the 16S rDNA profiles of soil bacteria obtained by denaturing gradient gel electrophoresis. 2016.
- Dec M, Puchalski A, Urban-Chmiel R, Wernicki A. 16S-ARDRA and MALDI-TOF mass spectrometry as tools for identification of Lactobacillus bacteria isolated from poultry. BMC Microbiology. 2016;16(1):1-16.
- Shah M. Amplified ribosomal DNA restriction analysis as a tool to characterize microbial community structure of activated sludge of common effluent treatment plant. International Journal of Environmental Bioremediation and Biogegradation. 2014;2:197-201.
- Horswell J, Cordiner SJ, Maas EW, Martin TM, Sutherland KBW, Speir TW, et al. Forensic comparison of soils by bacterial community DNA profiling. Journal of Forensic Science. 2002;47(2):350-3.
- Naknim V, Kutanan W, Lomthaisong K. Identifying the origin of forensic soil evidence using amplified ribosomal DNA restriction analysis of its bacterial community. J Nat Sci. 2016;15:115-28.
- Moreno LI, Mills D, Fetscher J, John-Williams K, Meadows-Jantz L, McCord B. The application of amplicon length heterogeneity PCR (LH-PCR) for monitoring the dynamics of soil microbial communities associated with cadaver decomposition. Journal of Microbiological Methods. 2011;84(3):388-93.
- Damaso N, Mendel J, Mendoza M, von Wettberg EJ, Narasimhan G, Mills D. Bioinformatics approach to assess the biogeographical patterns of soil communities: the utility for soil provenance. Journal of Forensic Sciences. 2018;63(4):1033-42.
- Pinkart H, Ringelberg D, Piceno Y, Macnaughton S, White D. Biochemical approaches to biomass measurements and community structure analysis. Manual of Environmental Microbiology (Hurst CJ, Crawford RL, Knudsen GR, McInerney MJ & Stentzenbach LD, eds). ASM Press, Washington, DC; 2002.
- Drenovsky RE, Steenwerth KL, Jackson LE, Scow KM. Land use and climatic factors structure regional patterns in soil microbial communities. Global Ecology and Biogeography. 2010;19(1):27-39.
- Veum KS, Lorenz T, Kremer RJ. Phospholipid fatty acid profiles of soils under variable handling and storage conditions. Agronomy Journal. 2019;111(3):1090-6.
- Ramsey PW, Rillig MC, Feris KP, Holben WE, Gannon JE. Choice of methods for soil microbial community analysis: PLFA maximizes power compared to CLPP and PCR-based approaches. Pedobiologia. 2006;50(3):275-80.
- Moter A, Göbel UB. Fluorescence in situ hybridization (FISH) for direct visualization of microorganisms. Journal of microbiological methods. 2000;41(2):85-112.
- Piwosz K, Mukherjee I, Salcher MM, Grujčić V, Šimek K. CARD-FISH in the sequencing era: opening a new universe of protistan ecology. Frontiers in Microbiology. 2021;12:640066.
- Ishii K, Mußmann M, MacGregor BJ, Amann R. An improved fluorescence in situ hybridization protocol for the identification of bacteria and archaea in marine sediments. FEMS Microbiology Ecology. 2004;50(3):203-13.
- Macdonald CA, Clark IM, Hirsch PR, Zhao F-J, McGrath SP. Development of a real-time PCR assay for detection and quantification of Rhizobium leguminosarum bacteria and discrimination between different biovars in zinc-contaminated soil. Applied and environmental microbiology. 2011;77(13):4626-33.
- Nuccio EE, Blazewicz SJ, Lafler M, Campbell AN, Kakouridis A, Kimbrel JA, et al. HT-SIP: A semi-automated Stable Isotope Probing pipeline identifies interactions in the hyphosphere of arbuscular mycorrhizal fungi. bioRxiv. 2022:2022.07. 01.498377.
- Li Z, Yao Q, Guo X, Crits-Christoph A, Mayes MA, IV WJH, et al. Genome-resolved proteomic stable isotope probing of soil microbial communities using 13CO2 and 13C-methanol. Frontiers in microbiology. 2019;10:2706.
- Young JM, Weyrich LS, Breen J, Macdonald LM, Cooper A. Predicting the origin of soil evidence: High throughput eukaryote sequencing and MIR spectroscopy applied to a crime scene scenario. Forensic Science International. 2015;251:22-31.
- Santillana E, Cordero JC, Alamilla F. Forensic soil analysis: case study of looting at a Roman-Visigothic burial vault. Soil in Criminal and Environmental Forensics: Springer; 2016. p. 45-60.
- Demanèche S, Schauser L, Dawson L, Franqueville L, Simonet P. Microbial soil community analyses for forensic science: application to a blind test. Forensic Science International. 2017;270:153-8.
- Cocking JH, Turley SR, Fofanov VY, Samuels-Crow K, Hungate B, Mau RL, et al. Forensic analysis of Soil Microbiomes: Linking Evidence to a Geographic Location. BioRxiv. 2020.
- Wang J, Zhang Q, Yuan M, Chu H, Shi Y. Developing a method for exploiting soil bacterial communities as evidence in environmental forensic investigations. Environmental Forensics. 2021;22(3-4):385-92.
- Handelsman J. Metagenomics: application of genomics to uncultured microorganisms. Microbiology and Molecular Biology Reviews. 2005;69(1):195-.
- Langille MG, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nature Biotechnology. 2013;31(9):814-21.
- Sa R, Cai L, Wu H, Yan J, Liu X, Hu R. Application of Metagenomics in Forensic Identification. Fa yi xue za zhi. 2017;33(4):397-400.
- Soliman T, Yang S-Y, Yamazaki T, Jenke-Kodama H. Profiling soil microbial communities with next-generation sequencing: the influence of DNA kit selection and technician technical expertise. PeerJ. 2017;5:e4178.
- Wei Y-j, Wu Y, Yan Y-z, Zou W, Xue J, Ma W-r, et al. High-throughput sequencing of microbial community diversity in soil, grapes, leaves, grape juice and wine of grapevine from China. PloS one. 2018;13(3):e0193097.
- Børsting C, Morling N. Next generation sequencing and its applications in forensic genetics. Forensic Science International: Genetics. 2015;18:78-89.
- Nkongolo K, Narendrula-Kotha R. Advances in monitoring soil microbial community dynamic and function. Journal of applied genetics. 2020;61(2):249-63.
- Young JM, Weyrich LS, Cooper A. Forensic soil DNA analysis using high-throughput sequencing: a comparison of four molecular markers. Forensic Science International: Genetics. 2014;13:176-84.
- Margenot AJ, Calderón FJ, Goyne KW, Dmukome FN, Parikh S. IR Spectroscopy, soil analysis applications. Encyclopedia of Spectroscopy and Spectrometry: Elsevier; 2016. p. 448-54.
- Young JM, Weyrich LS, Cooper A. High‐throughput sequencing of trace quantities of soil provides reproducible and discriminative fungal DNA profiles. Journal of Forensic Sciences. 2016;61(2):478-84.
- Hopkins JM. Forensic soil bacterial profiling using 16S rRNA gene sequencing and diverse statistics: Michigan State University; 2014.
- Jesmok EM. Adoption of next-generation 16s bacterial sequencing practices for the forensic analysis of soil: Michigan State University; 2015.
- Finley SJ, Benbow ME, Javan GT. Potential applications of soil microbial ecology and next-generation sequencing in criminal investigations. Applied Soil Ecology. 2015;88:69-78.
- Dunbar J, Ticknor LO, Kuske CR. Assessment of microbial diversity in four southwestern United States soils by 16S rRNA gene terminal restriction fragment analysis. Applied and Environmental Microbiology. 2000;66(7):2943-50.
- Osborne CA, Rees GN, Bernstein Y, Janssen PH. New threshold and confidence estimates for terminal restriction fragment length polymorphism analysis of complex bacterial communities. Applied and Environmental Microbiology. 2006;72(2):1270-8.
- Malik S, Beer M, Megharaj M, Naidu R. The use of molecular techniques to characterize the microbial communities in contaminated soil and water. Environment International. 2008;34(2):265-76.
- Muyzer G, De Waal EC, Uitterlinden AG. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Applied and Environmental Microbiology. 1993;59(3):695-700.
- Sun H, Deng SP, Raun WR. Bacterial community structure and diversity in a century-old manure-treated agroecosystem. Applied and Environmental Microbiology. 2004;70(10):5868-74.
- Gich FB, Amer E, Figueras JB, Abella CA, Balaguer MD, Poch M. Assessment of microbial community structure changes by amplified ribosomal DNA restriction analysis (ARDRA). International Microbiology. 2000;3(2):103-6.
- Spiegelman D, Whissell G, Greer CW. A survey of the methods for the characterization of microbial consortia and communities. Canadian Journal of Microbiology. 2005;51(5):355-86.
- Sardaro MLS, Perin LM, Bancalari E, Neviani E, Gatti M. Advancement in LH-PCR methodology for multiple microbial species detections in fermented foods. Food Microbiology. 2018;74:113-9.
- Roesch LF, Fulthorpe RR, Riva A, Casella G, Hadwin AK, Kent AD, et al. Pyrosequencing enumerates and contrasts soil microbial diversity. The ISME journal. 2007;1(4):283-90.
- Ari Ş, Arikan M. Next-generation sequencing: advantages, disadvantages, and future. Plant Omics: Trends and Applications: Springer; 2016. p. 109-35.
- Bayrak-Toydemir P, Wooderchak-Donahue W. Gene/Genome Mutation Detection and Testing. Pathobiology of Human Disease. 2014:3408-17.
- Wang J, Huang M, Wang Q, Sun Y, Zhao Y, Huang Y. LDPE microplastics significantly alter the temporal turnover of soil microbial communities. Science of the Total Environment. 2020;726:138682.
- Sun H, Wu Y, Zhou J, Bing H, Zhu H. Climate influences the alpine soil bacterial communities by regulating the vegetation and the soil properties along an altitudinal gradient in SW China. Catena. 2020;195:104727.
- Arias OV, Garrido A, Villeta M, Tarquis AM. Homogenisation of a soil properties map by principal component analysis to define index agricultural insurance policies. Geoderma. 2018;311:149-58.
- Ali A, Imran Ghani M, Li Y, Ding H, Meng H, Cheng Z. Hiseq base molecular characterization of soil microbial community, diversity structure, and predictive functional profiling in continuous cucumber planted soil affected by diverse cropping systems in an intensive greenhouse region of northern China. International Journal of Molecular Sciences. 2019;20(11):2619.
- Barba C, Folch A, Sanchez-Vila X, Martínez-Alonso M, Gaju N. Are dominant microbial sub-surface communities affected by water quality and soil characteristics? Journal of Environmental Management. 2019;237:332-43.
- Schöler A, Jacquiod S, Vestergaard G, Schulz S, Schloter M. Analysis of soil microbial communities based on amplicon sequencing of marker genes. Springer; 2017. p. 485-9.
- McDonald D, Price MN, Goodrich J, Nawrocki EP, DeSantis TZ, Probst A, et al. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. The ISME journal. 2012;6(3):610-8.
- Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Research. 2012;41(D1):D590-D6.
- Yilmaz P, Parfrey LW, Yarza P, Gerken J, Pruesse E, Quast C, et al. The SILVA and “all-species living tree project (LTP)” taxonomic frameworks. Nucleic Acids Research. 2014;42(D1):D643-D8.
- Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, et al. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Research. 2009;37(suppl_1):D141-D5.
- Sayers EW, Beck J, Bolton EE, Bourexis D, Brister JR, Canese K, et al. Database resources of the national center for biotechnology information. Nucleic Acids Research. 2021;49(D1):D10.
- Meyer F, Paarmann D, D’Souza M, Olson R, Glass EM, Kubal M, et al. The metagenomics RAST server–a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics. 2008;9(1):1-8.
- Meyer F, Bagchi S, Chaterji S, Gerlach W, Grama A, Harrison T, et al. MG-RAST version 4—lessons learned from a decade of low-budget ultra-high-throughput metagenome analysis. Briefings in Bioinformatics. 2019;20(4):1151-9.
- Chen I-MA, Markowitz VM, Chu K, Palaniappan K, Szeto E, Pillay M, et al. IMG/M: integrated genome and metagenome comparative data analysis system. Nucleic Acids Research. 2016:gkw929.
- Seshadri R, Kravitz SA, Smarr L, Gilna P, Frazier M. CAMERA: a community resource for metagenomics. PLoS Biology. 2007;5(3):e75.
- Mukherjee S, Stamatis D, Bertsch J, Ovchinnikova G, Sundaramurthi JC, Lee J, et al. Genomes OnLine Database (GOLD) v. 8: overview and updates. Nucleic Acids Research. 2021;49(D1):D723-D33.
- Huson DH, Auch AF, Qi J, Schuster SC. MEGAN analysis of metagenomic data. Genome Research. 2007;17(3):377-86.
- Choi J, Yang F, Stepanauskas R, Cardenas E, Garoutte A, Williams R, et al. RefSoil: A reference database of soil microbial genomes. BioRxiv. 2016:053397.
- Abdul Rahman NSN, Abdul Hamid NW, Nadarajah K. Effects of abiotic stress on soil microbiome. International Journal of Molecular Sciences. 2021;22(16):9036.
- Choi J, Yang F, Stepanauskas R, Cardenas E, Garoutte A, Williams R, et al. Strategies to improve reference databases for soil microbiomes. The ISME journal. 2017;11(4):829-34.
- Li W, Jiang L, Zhang Y, Teng D, Wang H, Wang J, et al. Structure and driving factors of the soil microbial community associated with Alhagi sparsifolia in an arid desert. Plos one. 2021;16(7):e0254065.
- Oliveira M, Amorim A. Microbial forensics: new breakthroughs and future prospects. Applied microbiology and biotechnology. 2018;102:10377-91.
- Habtom H, Pasternak Z, Matan O, Azulay C, Gafny R, Jurkevitch E. Applying microbial biogeography in soil forensics. Forensic Science International: Genetics. 2019;38:195-203.
Abbreviations
T-RFLP- Terminal Restriction Fragment Length Polymorphism
DGGE – Temperature Gradient Gel Electrophoresis
ARDRA – Amplified Ribosomal DNA Restriction Analysis
LH-PCR – Length Heterogeneity Polymerase Chain Reaction
PLFAs – Phospholipid Fatty Acids
FISH- Fluorescence In Situ Hybridization
SIP- Stable Isotope Probing
ICP-OES- Inductive Coupled Plasma Optical Emission Spectroscopy
MAGs – Metagenome-Assembled Genomes
NGS- Next Generation Sequencing
RDP- Ribosomal Database Project
NCBI- National Center for Biotechnology Information
MG-RAST- Metagenomic Rapid Annotations using Subsystems Technology
IMG/M- Integrated Microbial Genomes and Metagenomes
GOLD- Genomes OnLine Database
MEGAN- Metagenomic Analyzer
(Visited 1,117 times, 3 visits today)
This work is licensed under a Creative Commons Attribution 4.0 International License.





