Manuscript accepted on : 04-08-2022
Published online on: 27-08-2022
Plagiarism Check: Yes
Reviewed by: Dr. Ajar Nath Yadav
Second Review by: Dr. Olayinka O. Idris
Final Approval by: Dr. Ghulam Md Ashraf
Manubolu Harika Devi and Srinivas Munjam*
and Srinivas Munjam*
Department of Microbiology, Kakatiya University, Warangal-506009, Telangana state, India.
Corresponding Author E-mail: munjam17@gmail.com
DOI : http://dx.doi.org/10.13005/bbra/3022
ABSTRACT:
On account of its minimal expense and a high potential for change into energy-producing products, lignocellulosic biomass is the future of bioenergy and energy. Notwithstanding, their true capacity is restricted by the utilization of inefficient and unstable enzymes. Therefore, the present work aim to investigate the cellulose degradation potential of two isolated fungal species Aspergillus terreus and Trichoderma harzanium followed by bioethanol production from acid-thermal pre-treated rice straw (RS). The experiments were conducted in two phases. In phase-I of experiments, the isolated cellulose-degrading fungal species (Aspergillus terreus and Trichoderma harzanium) were employed for enzymatic hydrolysis of pretreated RS which was separated into two fractions namely: (Hydrolysate liquid) HL and (Residual pulp) RP. In phase-II of experiments, the enzymatically hydrolyzed substrate was subjected to yeast fermentation for bioethanol production. Using 18S rRNA sequencing, the microbial diversity study of the isolated species is covered in detail. The results revealed that the isolated fungal species A. terreus and T. harzanium resulted in 80 % of cellulose degradation. The highest bioethanol yield of 0.38 and 0.42 g/g of glucose was obtained from HL using Aspergillus terreus and Trichoderma harzanium treatment followed by yeast fermentation. The bioethanol yield of 0.17 g/g of cellulose was obtained from HL using both the fungal species.
KEYWORDS: Aspergillus terreus; Bioethanol; Cellulose; Rice straw; Trichoderma harzanium
Download this article as:| Copy the following to cite this article: Devi M. H, Munjam S. Bioethanol Production from Rice Straw and Cellulose Degradation using Aspergillus terreus and Trichoderma harzanium. Biosci Biotech Res Asia 2022;19(3). |
| Copy the following to cite this URL: Devi M. H, Munjam S. Bioethanol Production from Rice Straw and Cellulose Degradation using Aspergillus terreus and Trichoderma harzanium. Biosci Biotech Res Asia 2022;19(3). Available from: https://bit.ly/3KrfhFZ |
Introduction
Fossil fuels represent >80% of the total energy supplies in the world. The transportation area consumes around 29% of this, provoking worries about fossil product supplies, as well as its effect on the greenhouse effect, global warming, and environmental change. The current trend is towards the development of new techniques using renewable energy sources such as solar, wind, geothermal, and biomass1. Urbanization and modern development, then again, increased garbage production, which has social, environmental, and economic consequences. It can affect human health and populace security assuming it is misused, as well as expanding the deficiency of unrefined components, water, and fossil fuels2. It can likewise be monetarily disadvantageous on the off chance that the capital consumptions of new infrastructures are exceptionally high, and how much trash to be collected and treated surpasses the executive’s capacity. Accordingly, bio-based arrangements are being looked to further develop energy security, lessen petroleum product reliance, diminish CO2 emissions, and decrease waste generation. Because of its low-cost and high potential for transformation into energy-producing products, where lignocellulosic biomass is the future of bioenergy and fuels3. However, their potential is limited by the use of inefficient and unstable enzymes.
Cellulase is found in a variety of microorganisms, however, most of them are sluggish or unsuitable for industrial conversion. The development of an efficient process requires the selection of suitable microorganisms. The significant expense of cellulase enzymes is one of the main obstacles in the conversion of lignocelluloses to bioethanol. Cellulases have a great deal of potential in the modern world. Glucose produced from cellulosic substrate could be used as a substrate for additional fermentation or another interaction that produces significant-end products. Cellulases, like ethanol, methane, butanol, Single-cell protein (SCP), and others, have been utilized in waste-water treatment, food processing, detergent formulation, feed preparation, textile manufacture, and other industries for a very long time. An important stage in the real-time application is the screening of optimal enzymes with high catalytic activity and stability. Plant cellulose can be metabolized by a variety of bacteria and fungi4-5. The consolidated response of bacterial and fungal groups to cellulose in assorted soils, then again, stays unknown. Under both aerobic and anaerobic soil conditions, soil microorganisms can degrade cellulose. Therefore, bioethanol production could be a reasonable choice for actually using agricultural waste. As far as biomass accessibility, rice straw, corn straw, wheat straw, and sugarcane bagasse are the most widely recognized agricultural wastes. Besides, cellulases produced from different microbial sources contrast altogether with regards to strength, catalytic efficacy, and subsequently cellulose hydrolysis rate. A few techniques have been utilized to resolve this issue, including the quest for microbes with higher cellulase production capacities, the improvement of microbial cellulase synthesis through growth parameter optimization, and the advancement of effective cellulolytic systems through hereditary and protein engineering, all of which can fundamentally diminish the expense of the saccharification process6-7.
Stalks, stems, leaves, seedpods, husks, bagasse, and roots are a portion of the agricultural waste feedstocks with a ton of guarantee for biofuel generation8-9. Bioethanol is one of the most common biofuels produced using lignocellulosic biomass. Due to its minimal expense and a high potential for change into energy-producing products, lignocellulosic biomass is the future of bioenergy and fuels. In any case, their true capacity is restricted by the utilization of wasteful and unsteady fuels. Cellulase is found in a variety of microorganisms, however, most of them are sluggish or unsuitable for industrial conversion. As a result, researchers are looking into the many habitats of cellulase-producing bacteria to find effective and innovative cellulase or glycoside hydrolases. Bacteria and fungi are the two microbial species that are capable to degrade products of cellulose such as cellobiose, especially glucose. Actinobacteria and Streptomyces are the two dominant cellulolytic community groups found in soil. Because most species of the Aspergillus genus (Aspergillus niger) can manufacture cellulases under a variety of conditions, this genus can remove and overwhelm the enzyme industry and is notable for producing high-quality cellulase10-12.
After acid-thermal pretreatment, rice straw was separated into hydrolysate liquor (liquid fermentation) and residual pulp (solid-state fermentation) for bioethanol production. Therefore, the bioethanol production potential of the isolated fungal species for bioethanol production was evaluated using residual pulp and hydrolyzed liquid of rice straw as substrate. Using 18S rRNA gene sequencing analysis, the microbial diversity study of the isolated species is covered in detail. Therefore, the present work aims to look into the pretreated rice straw’s ability to produce bioethanol. The two isolated cellulose-degrading fungi such as Aspergillus terreus and Trichoderma harzanium are used to examine the effects of an acid-thermal pre-treatment followed by enzymatic hydrolysis.
Materials and Methods
Substrate collection
Dry rice straw (RS) of 1.5 kg was collected from the agricultural fields in the Warangal District of Telangana State, where the predominant cultivated crop is the paddy. RS Rice straw was cut into small pieces about 1 to 3 cm size using scissors before pretreatment. The raw RS was analyzed for total solids (TS), hemicellulose, volatile solids (VS), cellulose, lignin, nitrogen, carbon, hydrogen, and sulfur as per the standard methods.
Acid thermal pretreatment
Soaking the rice straw in the acidic solution followed by thermal pretreatment releases the monomeric units such as glucose from cellulose. Acid thermal pretreatment initially results in the breakdown of lignin bonds leading to the accessibility of cellulose for microorganisms to produce glucose which is one of the main feed components for the yeast to produce bioethanol. Therefore, acid thermal pretreatment was adopted in this study. Small pieces of 1 kg RS was soaked overnight in 7L of 1% H2SO4. The soaked RS in acid solution was then thermally pretreated in an autoclave at 1bar and 120oC for 20 minutes i.e., 1 cycle. The acid-thermal pretreated RS was pulpy and contained brownish liquid. The pretreated slurry was then filtered out to separate the hydrolysate liquid (HL) and the residual pulp (RP). Both the HL and RP were analyzed for physicochemical characteristics as per the standard analytical procedures mentioned in APHA13.
Isolation and identification of cellulose-degrading fungi
Isolation of cellulolytic fungi
The Comprehensive survey of different decomposing soils such as plant decomposing soil, vegetables decomposing soil, maize decomposing soil, paddy decomposing soil, wood decomposing soil, grass decomposing soil, flower decomposing soil, litter decomposing soil, sugarcane decomposing soil, dump yard decomposing soil and composting soil collected from different places of Warangal District, Telangana, India were analyzed for the presence of cellulose-degrading fungi. Each soil sample of 1.0 g was used for the serial dilution by using NaCl as diluent prepared with double distilled water up to 10-1-10-8 dilutions. Among these dilutions, 10-3 and 10-4 were taken for the isolation of different fungi by using the pour plate method containing potato dextrose agar. To accomplish uniform circulation of the sample, the media was tenderly poured into the petriplates and incubated at 27°C for 5 days. Antibiotics and rose bengal were used to reduce bacterial growth and fungal colonies. The fungi developed between the 3rd and 7th day were then isolated and identified by staining the isolated and cultured colonies with cotton blue and lactophenol by covering them with a glass coverslip. They were viewed under a light microscope with low power (10X) and high power (40X) objective lenses. A laboratory handbook for beginning mycology was used to identify morphological traits of fungus species14-15. The morphologically identified species were further confirmed by molecular identification using 18S rRNA gene sequencing analysis. The incidence, frequency, and abundance of individual fungi were also calculated in percentage using the following equations Eq. 1, Eq. 2, and Eq. 3.
Identification and screening of cellulolytic fungi by Congo red assay
The isolated fungi were screened for cellulolytic properties by cultivating on Czapek’s agar medium (Cellulose: 30 g; NaNO3: 3 g; KH2PO4.3H2O: 1.3 g; MgSO4.7 H2O: 0.5 g; KCl: 0.5 g; FeSO4. 7 H2O: 0.001 g; ZnSO4.7 H2O: 0.001 g; CuSO4.7 H2O: 0.0005 g; Yeast extract: 5 g; 20 g agar-agar and 1000 mL deionized water and pH around 6.3±0.2) by Petri plate method.At 30±2°C, all of the plates were incubated for 4 days. All Petri dishes were immersed with anaqueous solution of Congo red (1% percent w/v) for 15 minutes at the end of the incubation period, then Congo red solution was drained off. The plates were then subsequently treated by flooding them with 1 M NaCl for 15 minutes. The creation of a clear zone around the colonies is a cellulose degradation signal.
CMCase activity assay
Each fungal isolate was developed in a 150 mL Erlenmeyer flask containing 50 ml of basal salt medium modified with CMC (10 g/L) as the main carbon source to approve its cellulose-degrading limit. The pH of the medium was adjusted to 6.5. Every flask was inoculated with old PDA plate culture and refined for 5 days at 30±2°C. The crude enzyme was separated prior to being centrifuged for 10 minutes at 11,000 rpm. The enzyme activity was resolved to utilize Miller’s technique16, which comprised 0.2 mL of crude enzyme solution in addition to 1.8 mL of 1% carboxymethyl cellulose (CMC) in a 100 mM sodium phosphate cushion (pH 5.5). The reaction was then incubated in a water bath at 50°C for 25 minutes. The reaction was terminated by adding 3.0 mL of Dinitrosalysilic corrosive (DNS) reagent and setting the reagent tubes in a water bath at 100°C for 15 minutes to permit color advancement. Utilizing a standard curve, the OD was estimated at 575 nm, and the concentration of reducing sugars was assessed and computed as glucose.
Selection of strains for experimental studies
Among all the 50 selected fungal strains collected, 4403 colonies were isolated using different dilutions. Among 4403 colonies, 9 fungal species belonging to different genera were screened for cellulolytic properties. Based on the diameter of the cellulolytic zone that appeared on Czapek medium plates, two fungal species such as Aspergillus terreus strain and Trichoderma harzianum strain were selected for further studies (enzymatic hydrolysis and bioethanol production) and characterized phenotypically with the help of cultural and morphological characteristics. The morphological features were further confirmed by molecular identification using 18S rRNA sequencing analysis and the obtained sequences were deposited in NCBI.
Experimental design and operational procedure
The experiments in the present study were conducted in two phases. In phase I of experiments, the isolated cellulose-degrading fungal species (A. terreus and T. harzianum) were employed for enzymatic hydrolysis of pretreated RS which was separated into two fractions namely: HL and RP. In phase II of experiments, the enzymatically hydrolyzed substrate was subjected to yeast fermentation for bioethanol production. The design of the experiments is shown in Fig. 1.
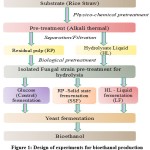 |
Figure 1: Design of experiments for bioethanol production. |
Enzymatic pretreatment using isolated cellulose-degrading fungal strains: Aspergillus terreus and Trichoderma harzianum
The hydrolysate liquor (HL) and residual pulp (RP) were subjected to enzymatic hydrolysis using the isolated A. terreus and T. harzianum fungal strains. The experiments were performed in batch mode in 250 mL Erlenmeyer flaskswith an effective volume of 200 mL from here on referred to as ‘Reactors’, labeled as R1 for A. terreus and R2 for T. harzianum to investigate the impact of the isolated strains on the degradation of cellulose. Two control reactors R1-Control using A. terreus and R2-Control using T. harzianum were also operated in parallel using cellulose as a carbon source. Therefore, a total of 6 reactors namely R1-Control; R1-HL; R1-RP; R2-Control; R2-HL, and R2-RP were operated in parallel for 5 days at mesophilic temperature (32±2oC). The reactors were fed with 150 mL of HL in R1 reactors and 150 mL of RP in R2 reactors. The R1 reactors were inoculated with 50 mL of A. terreus fungal strain in the form of a broth (fungal strains enriched in cellulose-degrading media) and an R2 reactor with 50 mL of T. harzianum strain respectively. In the control reactor, about 10 g of cellulose was used as feed which was considered control in the phase I experimental setup. The initial and final samples on day 1 and day 5 respectively were analyzed for cellulose, glucose, volatile fatty acids and soluble Chemical Oxygen Demand (COD) content to understand the performance of isolated fungal strains in degrading cellulose.
Yeast fermentation of enzymatically treated HL and RP for bioethanol production
In the second phase of experiments, the enzymatically hydrolyzed substrate was subjected to yeast fermentation for the production of bioethanol17. Similar to the phase I experiments, a set of 6 reactors were operated in parallel to understand the bioethanol production potential of the enzymatically treated substrate through yeast fermentation. Commercial grade yeast was purchased and enriched in a liquid medium containing yeast fermentation media. The supernatant liquid from both the reactors R1 and R2 was separated and fed to another set of reactors labeledR3 and R4. The supernatant from R1 (A. terreus treated) and R2 (T. harzianum treated) reactors were fed to R3 and R4 reactors respectively. The reactors (R1 and R2) containing HL and RP respectively were subjected to liquid and solid-state fermentation. The reactors R3 (control; HL and RP) and R4 (control; HL and RP) were inoculated with 50 mL of enriched yeast. The reactors were operated for 4 days to observe the bioethanol production from RS after enzymatic pretreatment. The initial and final samples on day 1 and day 5 respectively were analyzed for pH and bioethanol production to understand the efficiency of enzymatic pretreatment followed by yeast fermentation in bioethanol production.
Results and Discussion
Characteristics of the raw substrate and impact of acid thermal pretreatment
The characteristics of rice straw before and after acid thermal pretreatment is tabulated and the average values are presented in Table 1.0. As can be seen from Table 1.0. the cellulose, hemicellulose, and lignin content in the acid-thermally pre-treated RS decreased from 34–24%; 23–13%, and 17–7% respectively. The carbon content in raw RS was found to be 41% suggesting that the RS is suitable for biofuel production. However, the presence of lignin hinders the easy accessibility of the cellulose and hemicellulose to the microorganisms for their conversion to its monomers for bioethanol production. The Chemical Oxygen Demand was 45 g/L in HL and 40 g/L in RP indicating that the pre-treated substrate is organic-rich. The VFA in HL and RP was 12 g/L and 7 g/L respectively. Overall, the acid-thermal pre-treatment has positively impacted the substrate in increasing the accessible organic matter in the form of soluble Chemical oxygen Demand, Volatile Fatty acids, and Volatile Solids which are essential food to microorganisms for bioethanol production.
Table 1. Raw and pre-treated slurry characteristics of the substrates.
| Parameter | Unit | Raw Rice Straw (RS) | Acid-thermal treated RS | Hydrolysate liquor (HL) | Residual pulp (RP) |
| Total Solids (TS) | % | 94 | 15 | 9 | 75 |
| Volatile Solids (VS) | % | 85 | 13 | 8 | 70 |
| Cellulose | % | 34 | 24 | 18 | 32 |
| Hemicellulose | % | 23 | 13 | 6 | 23 |
| Lignin | % | 17 | 7 | 2.5 | 15 |
| COD | g/L | ND | ND | 110 | 140 |
| VFA | g/L | ND | ND | 12 | 7 |
| sCOD | g/L | ND | 85 | 45 | 40 |
| Carbon (C) | % | 41 | 36 | ND* | ND* |
| Hydrogen (H) | % | 6.7 | 4.5 | ND* | ND* |
| Nitrogen (N) | % | 0.2 | 0.5 | ND* | ND* |
| Sulfur (S) | % | 0.2 | 0.1 | ND* | ND* |
Activity test of isolated cellulolytic fungal strains
Among all the 50 selected fungal strains collected, 4403 colonies were isolated using different dilutions. The isolated microorganisms belonged to fungal species which represented 9 genera (Alternaria, Aspergillus, Cladosporium, Curvularia, Fusarium, Mucor, Rhizopus, Penicillium, and Trichoderma) among the isolates. Among 4403 colonies, 9 fungal species belonging to different genera were screened to be positive for cellulolytic properties. The growth region of the cellulolytic activity of the isolated fungal strains, stained on Congo red in the Czapek medium plates is shown in Fig. 2. Based on the diameter of the cellulolytic zone that appeared on czapek medium plates, two fungal species such as A. terreus and T. harzianum were selected for further studies and characterized phenotypically with the help of cultural and morphological characteristics. The diagnostic features were further confirmed by molecular identification using 18S rRNA sequencing analysis and the obtained sequences were deposited in NCBI with accession numbers MZ617278 and MZ620641.
 |
Figure 2: Plates showing the growth of cellulolytic activity of the isolated cellulose-degrading fungi on Czapek medium. Click here to view figure |
Table 2: Screening of cellulolytic activity.
| Isolated Fungal species | Activity for cellulose degradation | Diameter of the cellulolytic zone (cm) |
| Alternaria | ++ | 2.1 |
| Aspergillus | +++ | 4.5 |
| Cladosporium | + | 1.5 |
| Curvularia | ++ | 2.5 |
| Fusarium | + | 0.8 |
| Mucor | ++ | 1.9 |
| Rhizopus | ++ | 2.8 |
| Penicillium | + | 1.1 |
| Trichoderma | +++ | 4.2 |
| Moderate growth: +; Good growth: ++; Maximum growth: +++ | ||
Phylogenetic identification and genetic characterization of the isolated fungal strains using 18S rRNA sequencing
The 18S rRNA sequencing of the isolated fungal strains from soil samples was carried out and labeled as HJSMKU-01and HJSMKU-02 respectively. The genetic characterization and the phylogenetic tree of the two isolated fungal species are shown in Fig. 3. Fig. 3(A) Fig. 3(B). The phylogenetic tree of both the isolated strains confirms the fungal strains isolated from the soil as A. terreus and T. harzanium respectively. These two fungal strains were known to degrade cellulosic material which is evident from the activity test and was capable of producing bioethanol. The gene bank submission number of both the species is SUB10066977 and SUB10067688 while the NCBI numbers are allocated for the isolated species from soil which were identified as A. terreus and T. harzanium are MZ617278 and MZ620641 respectively.
 |
Figure 3: Phylogenetic tree of the isolated fungal strains. |
Performance evaluation of enzymatic pretreatment of the substrate using isolated strains
The performance evaluation of enzymatic pretreatment of RS (HL and RP) and synthetic cellulose (carbon source) using A. terreus and T. harzanium in terms of cellulose, glucose, Volatile Fatty Acids, and soluble Chemical Oxygen Demand profiles is shown in Fig. 4. As can be seen from Fig. 4(a) that there is a considerable decrease in the cellulose content (%) in the substrate after enzymatic pretreatment. For instance, in control reactors R1 and R2 with synthetic cellulose as substrate, about 80 % of cellulose reduction was observed i.e., from 10 g to 8 g. The 80% reduction in cellulose has resulted in 6 g/L of glucose conversion using both A. terreus and T. harzanium.Interestingly, the performance of A. terreuson the reduction of cellulose with HL and RP as substrate was 92% and 80% respectively. Improved cellulose reduction using HL in comparison to RP could be attributed to the fact that HL was in liquid form while RP was solid which might have decreased the accessibility of cellulose to the fungi. Similarly, the performance of T. harzaniumon the reduction of cellulose with HL and RP as substrate was 93% and 82% respectively. A marginal difference in the cellulose reduction efficiency was observed in both species. However, there exists a difference in the formation of glucose from cellulose during enzyme hydrolysis i.e., 12.15 g/L and 16 g/L using A. terreus and 10.8 g/L and 21.6 g/L using T. harzanium. The main mechanism behind the conversion of cellulose to glucose using cellulose-degrading fungi is that the cellulose is a polymer consisting of monomeric glucose units linked by β-1, 4- glycosidic bonds in a linear mode which can only be accessible to microorganisms upon pretreatment. The breakdown of cellulose to glucose is the result of the coordinated activity of endocellulases, exocellulases (cellobiohydrolases, glucanohydrolases), and beta-glucosidases. Endocellulases randomly hydrolyze the internal glycosidic bonds resulting in a rapid decrease in polymer length and a gradual increase in the reducing sugar concentration17-18. Exocellulases hydrolyze cellulose polymers by releasing mostly cellobiose from either the reducing or non-reducing ends, resulting in rapid reducing sugar release with little change in polymer length. On cellulose, endocellulases and exocellulases work together to form cellooligosaccharides and cellobiose, which are then broken into glucose by beta-glucosidase5-6. Therefore, cellulose which is the most abundantly available organic matter in agriculture wastes is a valuable raw material that can be used as feed or can be converted to fuel and chemicals.
The enzymatic pretreatment using A. terreus and T. harzanium on the soluble Chemical oxygen Demand and Volatile Fatty acids profile was illustrated in Fig. 4(b). VFAs are the short-chain carboxylic acids mainly comprising acetic acid, propionic acid, and butyric acid which can also be converted into bioethanol19. The sCOD which is the completed biodegradable fraction of COD has improved significantly after enzymatic hydrolysis in reactors operated with RP because the cellulose content in the RP was relatively higher than that of HL in the reactors. Therefore, the activity of A. terreus and T. harzanium was superior in the reactors with cellulose-rich substrate i.e., RP on the other hand, HL also contained cellulose which resulted in the sCOD but was relatively lower than the HL reactors. The percentage increase in sCOD in reactors fed with HL was 77% using A. terreus and 69% using T. harzanium whereas in reactors with RP, it was 55% and 68% using A. terreus and T. harzanium respectively. This illustrates that the activity of T. harzanium on RP was more while it was HL for A. terreus. In contrast to the sCOD profile, the VFAs were more in reactors fed with HL compared to RP.
 |
Figure 4: Impact of acid thermal pretreatment followed by enzymatic hydrolysis on the degradation and conversion of cellulose to glucose, sCOD, and VFA |
Bioethanol generation potential of treated RS using solid-state and liquid state yeast fermentation
The performance evaluation of enzymatic pretreatment of RS (HL and RP) and synthetic cellulose (carbon source) using A. terreus and T. harzanium in terms of cellulose, glucose, VFA, and sCOD profiles is shown in Fig. 5. As can be seen from Fig. 5(a) theglucose formation and reduction are inversely related, i.e., in control reactors the formation of glucose is relatively lower compared to HL and RP reactors but the conversion of glucose to bioethanol using yeast through fermentation process is more in control reactors followed by HL and RP reactors. This could be due to two main reasons; one is the substrate inhibition and the other is the presence/formation of other intermediates that might have hindered the process. The pH in the reactors after A. terreus enzyme treatment was 3.8, 4.8, and 4.5 in control, HL, and RP reactors respectively whereas the pH is 5.0, 4.9, and 4.8 in T. harzanium reactors. However, after yeast fermentation, a reduction in pH was observed in all the reactors which areindicative of the production of bioethanol. As bioethanol can be produced at a pH range of 3.5 to 6.0 depending on the type of substrate and inoculum used. Comparable bioethanol production was observed in control, HL, and RP reactors. However, the T. harzanium treated RP of rice straw has resulted in more bioethanol of 5.4 g/L whereas it is 4.7 g/L in the case of A. terreus. Interestingly, the performance of A. terreus and T. harzanium in the reactors with HL as the substrate is the same with 4.5 g/L of bioethanol production. The control reactors with synthetic carbon sources have resulted in higher bioethanol production of 5.0 g/L and 5.5 g/L in both the cases of A. terreus and T. harzanium treatment which is obvious because the substrate was pure, unlike the other reactors which were fed to HL and RP of pretreated RS.
The bioethanol yield obtained from the reactors using yeast fermentation of the acid thermal followed by enzymatically treated fractions of RS (HL and RP) is reflected in Fig.5(b). As can be seen from Fig. 1.5(b) that the bioethanol yield of 0.44, 0.38, and 0.28 g/g of glucose was obtained from reactors R3-control, R3-HL, and R3-RP whereas it was 0.49, 0.42, and 0.25 g/g of glucose in reactors R4-control, R4-HL, and R4-RP. The bioethanol yield of 0.26, 0.17, and 0.10 g/g of cellulose were obtained from reactors R3-control, R3-HL, and R3-RP whereas was 0.29, 0.17, and 0.11 g/g of cellulose in reactors R4-control, R4-HL, and R4-RP. In summary, it can be stated that the bioethanol production using synthetic cellulose as substrate and the application of acid-thermal followed by enzymatic pretreatment is superior compared to that of real substrates such as the fraction of RS i.e., HL and RP.
 |
Figure 5: Performance evaluation of yeast fermentation using enzymatically acid thermal followed by enzymatically treated HL and RP. |
Conclusion
Valorization of agriculture wastes for bioethanol production by overcoming the substrate limitations for efficient degradation of cellulose and hemicellulose in the presence of recalcitrant lignin through the development of cellulose-degrading microorganisms is one of the promising approaches as the conversion of lignocelluloses to bioethanol using high-cost commercial cellulase enzyme makes the entire process expensive. Therefore, this study demonstrated the isolation, identification, and of two potential cellulose-degrading fungal species isolated from soil potential and tested for cellulose degradation of RS followed by bioethanol production via yeast fermentation. The 18S rRNA gene sequencing study showed that a total of 19 species capable of degrading cellulose were isolated and among them, about 9 species were found to be good for cellulose degradation. Among the 9 species, two species namely Aspergillus terreus and Trichoderma harzanium have shown maximum growth for cellulose degradation which were used as potential organisms in the study. Bioethanol production from the HL and RP of the acid-thermal followed by enzymatic hydrolysis pre-treatment of RS was used as feed. The isolated fungal species A. terreus and T. harzanium resulted in the degradation of cellulose by 80%. Yeast fermentation of hydrolyzed HL and RP resulted in the highest bioethanol yield of 0.38 and 0.42 g/g of glucose from HL using A. terreus and T. harzanium treatment. The bioethanol yield of 0.17 g/g of cellulose was obtained from HL using both the fungal species upon yeast fermentation.
Acknowledgment
The authors are grateful to the Head, Department of Microbiology, Kakatiya University, Warangal for the continuous encouragement and support to carry out this research work. The authors also thank the technical staff for their valuable services to perform the work.
Conflict of Interest
There is no conflict of interest.
References
- Owusu PA., Asumadu-Sarkodie S. A review of renewable energy sources, sustainability issues and climate change mitigation. Cogent Eng. 2016; 3(1):1167990.
CrossRef - McMichael AJ., Campbell-Lendrum, DH., Corvalán, CF., Ebi KL., Githeko A., Scheraga JD.,Woodward A. Climate change and human health: risks and responses. World Health Organization 2003.
- Anwar Z., Gulfraz M., Irshad M. Agro-industrial lignocellulosic biomass a key to unlock the future bio-energy: a brief review. Radiat. Res. Appl. Sci. 2014; 7(2):163-73.
CrossRef - Thomas L., Parameswaran B., Pandey A. Hydrolysis of pretreated rice straw by an enzyme cocktail comprising acidic xylanase from Aspergillus for bioethanol production, Renew. Energy. 2016; 98: 9-15.
CrossRef - Li DC., Papageorgiou AC. Cellulases from thermophilic fungi: recent insights and biotechnological potential. Fungi in Extreme Environments: Ecological Role and Biotechnological Significance. 2019; 395-417.
CrossRef - Harris PV., Welner D., McFarland KC., Re E., Navarro Poulsen JC., Brown K., Lo Leggio L. Stimulation of lignocellulosic biomass hydrolysis by proteins of glycoside hydrolase family 61: structure and function of a large, enigmatic family. Biochemistry, 2010; 49(15): 3305-3316.
CrossRef - Saranraj P., Stella D., Reetha D. Microbial cellulases and its applictions. j. biochem. biotechnol. 2012; 1:1-2.
- Braide (Dr) Wesley., Kanu IA., Solomon Oranusi., Samuel Adeniyi Adeleye. Production of bioethanol from agricultural waste. Journal of Fundamental and Applied Sciences. 2016; 8(2): 372.
CrossRef - Singhania RR., Adsul M., Pandey A., Patel AK. Cellulases. In Current developments in biotechnology and bioengineering 2017; (pp. 73-101). Elsevier.
CrossRef - Lejeune R., Baron G. Effect of agitation on growth and enzyme production of Trichoderma reesei in batch fermentation, Microbiol. Biotechnol. 1995; 43(2): 249-258.
CrossRef - Purwanto, D. Ibrahim, H. Sudrajat, Effect of agitation speed on morphological changes in Aspergillus niger hyphae during production of tannase, World J. Chem. 2009; 4(1): 34-38.
- Darwesh OM., El-Maraghy SH., Abdel-Rahman HM., Zaghloul RA. Improvement of paper wastes conversion to bioethanol using novel cellulose degrading fungal isolate. Fuel. 2020; 262:116518.
CrossRef - APHA-AWWA-WPCF. Standard Methods for the Examination of Water and Wastewater. American Public Health Association, Washington, D.C, 1998.
- Gilman Henry, Haubein AH. The Quantitative Analysis of Alkyllithium Compounds1.” Journal of the American Chemical Society. 1944; 66(9): 1515-1516.
CrossRef - Smith Geoffrey L., Bernard Moss. Infectious poxvirus vectors have capacity for at least 25 000 base pairs of foreign DNA. 1983; 25(1): 21-28.
CrossRef - Miller GL. Use of Dinitrosalicylic Acid Reagent for Determination of Reducing Sugar. Analytical Chemistry. 1959; 31(3): 426–428.
CrossRef - Mihajlovski K., Radovanovi, Z., Carevi M., Dimitrijevi-Brankovi S. Valorization of damaged rice grains: optimization of bioethanol production by waste brewer’s yeast using an amylolytic potential from the Paenibacillus chitinolyticus CKS1, Fuel. 2018; 224: 591-599.
CrossRef - Pietrzak W., Kawa-Rygielska J. Ethanol fermentation of waste bread using granular starch hydrolyzing enzyme: effect of raw material pretreatment. Fuel. 2014; 134:250-6.
CrossRef - Ward NL., Challacombe JF., Janssen PH., Henrissat B., Coutinho PM., Wu M., Xie G., Haft DH., Sait M., Badger J., Barabote RD. Three genomes from the phylum Acidobacteria provide insight into the lifestyles of these microorganisms in soils. Appl Environ Microbiol. 2009; 75(7): 2046-56.
CrossRef

This work is licensed under a Creative Commons Attribution 4.0 International License.






