Manuscript accepted on : 06-11-2021
Published online on: 19-11-2021
Plagiarism Check: Yes
Reviewed by: Dr. Nilay Nandi
Second Review by: Dr. Ahmed Lafi
Final Approval by: Dr. Hifzur siddique
Shigellosis and Development of Multiple Antimicrobial Resistance Mechanisms of Shigella Spp.
SK Tousif Ahamed and Nabanita Giri*
Department of Microbiology, Assistant professor, Acharya Prafulla Chandra College, New Barrackpore, West Bengal,India,700131.
Corresponding Author E-mail: nabanita@apccollege.ac.in
DOI : http://dx.doi.org/10.13005/bbra/2953
ABSTRACT:
Shigellosis is a serious public health issue. Millions of people suffer from this deadly food and water borne disease each year. The main manifestations of affected persons are bloody diarrhea with excessive dehydration. The causative agent of this disease is the bacteria Shigella spp. which has four serogroups. Though Shigella flexneri and Shigella dysenteriae are the dominant serogroups in developing countries, reports of other serogroups, namely Shigella boydii and Shigella sonnei, in the food contaminations are available. There are seasonal variations of Shigella infection throughout the world. In Asian subcontinent, monsoon and post monsoon times are the ideal for infection. The transmission of the bacteria in human is usually caused by feco-oral route or by contaminated food and water. There are several groups of antibiotics like foscomycin, macrolide, amiglycoside, tetracycline etc. which were used before. But they are now become useless as Shigella spp. is getting resistant against those drugs. The quinolone groups of antibiotics like ciprofloxacin, ofloxacin, norflxacin, ceftriaxone etc. are the important drugs for the cure of the disease shigellosis but prevalence of drug resistant strains of Shigella spp. against those drugs are a great concern nowadays. The occurrence of plasmid mediated quinolone resistance genes (PMQR), efflux pump proteins and effective mutations at drug binding region of gyrA etc. are the major mechanisms for the development of drug resistance.
KEYWORDS: Drug resistance; Efflux proteins; Pathogenicity; Serogroup; Virulence factors
Download this article as:| Copy the following to cite this article: Ahamed S. K. T, Giri N. Shigellosis and Development of Multiple Antimicrobial Resistance Mechanisms of Shigella Spp. Biosci Biotech Res Asia 2021;18(4). |
| Copy the following to cite this URL: Ahamed S. K. T, Giri N. Shigellosis and Development of Multiple Antimicrobial Resistance Mechanisms of Shigella Spp. Biosci Biotech Res Asia 2021;18(4). Available from: https://bit.ly/3Cxms9O |
Introduction
Shigellosis is an infectious gastrointestinal disease. It is caused by Shigella serogroups of four types. They are responsible for the diarrhea in children and adults with morbidity and mortality specially in developing countries1. Infection due to the Shigella spp. were estimated to be about 170 million per year with about 1 million deaths in developing countries, 69% of them are children under 5 years2. Although this number has decreased but it has remained one of the important heath threat around the world. The children and young adults of developing countries of South Asia, Southeast Asia and Africa are mostly affected by Shigella infections3. A large number of people suffer from gastroenteritis in Bangladesh every year4. The burden of shigellosis is mainly associated with the poor sanitization, poor healthcare, contaminated water and food. Infection is common by feco oral route5. The genous Shigella having four serogroups (Shigella flexneri, Shigella dysenteriae, Shigella sonnei and Shigella boydii). The occurrence of most common Shigella serogroups differs geographically. In underdeveloped countries Shigella dysenteriae and Shigella flexneri are common and generally spread by environmental surface water sources6. On the other hand, Shigella sonnei and Shigella boydii are reported mostly in industrialized countries and usually spread through food sources. Antimicrobial and oral rehydration therapy are recommended to treat this disease. World Health Organization (WHO) recommended different antibiotic such as ceftriaxone, azithromycin, ciprofloxacin, pivmeccillinam, for the treatment of shigellosis but the indiscriminate use of antibiotics results in the emergence of multidrug (MDR) resistant Shigella spp. which makes shigellosis a global threat7. Particularly resistance to quinolones and cephalosporin makes the situation more challenging8. Various antimicrobial resistance mechanism in Shigella spp. have been described by the researcher such as extrusion of drug by activating efflux pump, decrease in cellular permeability, overexpression of drug-modifying and inactivating enzymes or target modification by mutation9. Thus the recurrent changes in antimicrobial resistance pattern among the Shigella spp. poses the complication in recommending standard drug for effective treatment of shigellosis. The current study was done to review the emergence of antimicrobial resistance among the Shigella spp.
Shigellosis as an infectious acute dysentery
Infectious diseases are a major burden and the common cause of death worldwide. More than millions of people die each year in developing countries due to this disease among them most of are the children2. Dehydration by excessive fluid and electrolyte loss in stool results in acute dysentery which is the main reason of death. In developing countries diarrheal disease is not only a major health issue but also a risk factor to the travelers who travel to these countries. Treatment and preventive measures including fluid replacement and improvement of hygiene decrease the mortality rate from about 5 million to approximately 1.5 million deaths throughout the past two decades10. However, increased frequency of multi-drug resistant bacteria posed the problem more challenging7,8,11. Vibrio cholerae, multiple serovars of Salmonella enterica, different serotypes of Shigella, Campylobacter spp. and enteropathogenic Escherichia coli strains including Escherichia coli (ETEC) with the enteric viruses including rota viruse are the most important organisms that are frequently associated with diarrhea12. Acute watery diarrhea, dysentery and persistent diarrhea are the different types produced by those pathogens. Among the enteric pathogens, Shigella spp. is the most pathogenic organism that causes bacillary dysentery or shigellosis. Infection occurs by feco-oral route with the contaminated food and water5. Shigellosis is manifested by fever, abdominal cramps including the passage of loose stools mixed with blood and mucous. The annual number of shigellosis was estimated to be about 170 million episodes at the end of the last century with about 1 million deaths in developing countries2. Global projections suggest that there are 165 million episodes of shigellosis per year, of which 99 percent occurs in less than 5 years of age13. Although the number has reduced, shigellosis still remains one of the most important endemic diseases in the world 2.
Overview of Shigella spp.
Kyoshi Shiga in 1896 first isolated the Shigella dysentery at the Kitasato institute, Japan. He isolated a Gram negative bacillus from stool sample which showed positive agglutination reaction against sera of patient recovering from acute dysentery. Primarily he proposed the name of this organism as Bacillus dysentericus in 1897, later he changed it to Bacillus dysenteriae in 189814. During his research he described the production of toxic factor by the organism that now known as Shiga toxin. Few years later German microbiologist Kurse (1900) isolated two similar organisms from patients’ stool samples but they were serologically different from Bacillary dysenteriae. Over the two decades additional three groups of related organisms were identified and taxonomically placed in the Shigella and named as Shigella dysenteriae, Shigella flexneri, Shigella boydii and Shigella sonnei to honor the Shiga, Flexner, Boyd and Sonne. The genus Shigella was first termed in the 1930 edition of Bargey’s Manual of Determinative Bacteriology 15.Shigella belongs to Enterobacteriaceae which are small, nocapsulated, non-motile, non sporulating facultative anaerobic Gram negative bacilli. Genus Shigella has four major groups with several serotypes. They are – Shigella dysenteriae (13 serotype), Shigella flexneri (15 serotype), Shigella boydii (18 serotypes), Shigella sonnei (One serotype)16.
Shigella can also be grouped into different serogroups including A, B, C and D depending on the basis of common specific polysaccharide antigens present on the cell surface. Whole genome sequence of E. coli and all four Shigella spp. revealed that they share a common DNA backbone of approximately 3.9 Mb, interrupted by E. coli specific and Shigella specific sequences. S. dysenteraie and S. flexneri are the most common serotypes found in developing countries. S. dysenteraie type 1 usually cause epidemic and pandemic bacillary dysentery with severe form of disease and produce potential lethal complications due to the production of Shiga toxin. In Latin America, Asia and Europe S. dysentariae type 1 causes epidemic dysentery in 1960. Asian countries such as Bangladesh (1972-78, 2003), Sri Lanka (1976), Maldives (1982), Nepal (1984-85) and Bhutan (1984-85) reports the dysentery epidemic. In India epidemics due to Shigella infections were reported from different parts of the country including eastern India (1984), Andaman and Nicobar Island (1986), Chandigarh (2003), Vellore (1972-73,1997-2001) , West Bengal (2002-03) in different times2. Frequent and occasional Shigella infections were reported during summer and winter from Israel 17. Emergence of multidrug-resistance is the major problem in developing countries. In India especially in Kolkata S. dysenteraie type 1 became resistant to common antibiotics which results in high restriction in treatment18. Occasionally S. dysenteraie type 2 infections were reported and sometimes exceeded type 1 in the frequency of isolation. Other serotypes were encountered rarely among the diarrheal patient’s sample.
In developing countries mostly S. flexneri causes endemic. The serotypes 1b, 2a, 3a, 4a, and 6 were encountered in industrialized countries19. Several uncommon serotypes and sub-serotypes of S. flexneri were also isolated from different countries like Bangladesh, Egypt and Russia20,21. Outbreak of S. flexneri 2a in India as well as in Taiwan and United State were also reported22,23.
Outbreak due to S. sonnei is very rare in developing countries. There is no report of S. sonnei outbreak from India. Although it cause sequential outbreak but the mixture of genes with different origin found on S. sonnei plasmid due to the insertion of elements and number of other open reading frame. Shigella enterotoxin 2 prevalent in S. sonnei and a virulence gene sat has also been reported in this serotype24.
boydii is mainly reported from Indian subcontinent and which is least prevalent in developing countries25. S. boydii is mainly spread throughout the World by travelers. It is the second dominant serogroups in India followed by S. flexneri. New serovar of S. boydii has been reported from Bangladesh in 200526.
Across the world the incidence of Shigella spp. depend upon the seasonal pattern. During summer or early autumn Shigella infection were most common in US and Europe. Kagalwalla et.al in 1992 reported the most of the Shigella serotypes were isolated during April-May in Saudi Arabia. Increase incidence of Shigella was reported form Bangladesh during post-monsoon season of August-November and pre-monsoon from the month of April – May of each year. In India specially in Kolkata Shigella infection was prevalent throughout the year with high isolation rate during the summer and early monsoon months8,27,28. Shigella spp. were detected more in June to July, i.e. during rainy season in Laos and Guatemala. In Hungary July to September was the peak period for the isolation of S. flexneri and S. sonnei29.
 |
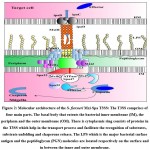
Figure 1: Mechanism of Shigella pathogenesis: S. flexneri encounters host macrophages after crossing the epithelial barrier (EC) and entry into M cells. |
Transmission and epidemiology
Shigellosis is endemic throughout the world, although one century is past after K. Shiga’s remarkable discovery but still it remains a global health problem. World Health Organization (WHO) estimated that 164.7 million cases of shigellosis occurred per year of which 1.1 million cases result in death in developing countries due to the poor hygiene, limited access of clean water and malnutrition. Shigella required only 10 -100 organisms to cause infection30. It is transmitted by feco-oral route or by the contaminated food and water30,31. Transmission by house flies has also been documented32. Outbreaks due do the Shigella spp. have been reported from different parts of the world as well as from India. In 2007 an outbreak due to the S. flexneri in West Bengal has been reported which affect more than 461 people33. Another foodborne outbreak of S. sonnei infection was reported from Pakaplole village, South 24 Parganas, India which affect more than 25 people in 201834. In Iran an outbreak due to the S. flexneri 2a affect more than 700 prisoners in 200735,36,37. Similarly in US an outbreak of shigellosis in 2006 was associated with the tomatoes23. MDR Shigella spp. outbreak has been reported from the Children Welfare Institute, China in 20154.
Antibiotic resistance profile of Shigella spp.
There are number of antibiotics recommended by WHO for the treatment of shigellosis which reduce the number of deaths per year but the extensive use of them led to an increase in isolation of multidrug-resistant Shigella spp. in several countries 7,11,19,38,39,40. Still shigellosis remains one of the most important endemic diseases in the world. In recent day’s treatment with ciprofloxacin [a fluoroquinolone (FQ)] or one of the three second-line antibiotics, pivmecillinam, azithromycin and ceftriaxone (a third-generation cephalosporin), have been recommended. However reports are available of FQ-resistant Shigella isolates from India and other Asian countries which become a serious concern to treat shigellosis41,42,43. Antibiotic resistance genes may be located in plasmid, chromosome and mobile genetic elements like integrons and transposons of the bacteria (Table 1).Mutation in the drug target or genes associated with efflux system also cause the drug resistance (Table 2).
Centre for Disease Control and prevention (CDC) (https://wwwn.cdc.gov/narmsnow/) reports that year wise antibiotic resistant pattern among the isolated Shigella spp. is a serious threat (Fig.3). There are various mechanisms which result in resistance to antibiotics to the Shigella spp. including extrusion of drugs by activating efflux pump, decrease in cellular permeability, modifications of target sites by mutations and over expression of enzymes which modify or inactivate the drugs44,45. The possible mechanisms are listed below-
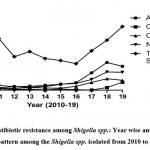 |
Figure 3: Antibiotic resistance among Shigella spp.: Year wise antibiotic resistant pattern among the Shigella spp. isolated from 2010 to 2019. |
Function of outer membrane permeability
Cell wall of microorganisms is served as first barrier for penetration of antimicrobial drug into the cell. Some modification or changes in the membrane result in porin loss which increase the minimum inhibitory concentration (MIC) to the antibiotics46. Most of the antibiotics which are used to treat Shigella infections should be able to penetrate through cell membrane to reach intercellular accumulation and target site. Quinolone group of antibiotics such as nalidixic acid, ofloxacin, ciprofloxacin interfere with DNA gyrase and topoisomerase IV to inhibit DNA replication. Aminoglycoside antibiotics including streptomycin and spectinomycin bind with ribosomal subunits and inhibit protein synthesis.β-lactam antibiotics such as cephalosporin and penicillin target the penicillin binding protein and inhibit the cell wall biosynthesis. Mutations or absence of a ~39 kDa porin in the membrane of Gram negative bacteria as for example Shigella spp. slowdown the penetration of β-lactam (Aztreonam and Dianionic moxalactam) and hydrophilic antibiotics such as penicillin and piperacillin44,46. Three mutant strain of S. dysenteriae isolated in India showed resistance to imipenem and the study reported that the resistance towards the imipenem is associated with the permeability of outer membrane proteins. It is also reported that the resistance to colicin E2 among the S. flexneri strains associate with LPSs of the outer membrane47.
Role of efflux system
Activation of efflux pump plays an important role in antibiotic resistance phenotype of Shigella spp. to expel the toxic compounds from the cells. Efflux system can be grouped into five families such as- the major facilitator super family, small multidrug resistance family, resistance-nodulation-division family, multidrug extrusion family and ATP binding cassette family48. In Shigella spp. a triplet complex namely AcrAB-Tol C, belonging to resistance-nodulation-division family of efflux system reduce the level of quinolone accumulation inside the cells which results in the resistance to quinolones49. It is also reported that drug efflux pump such as marA, tolC, YdhE and mdfA also confer resistance to quinolones45.
Resistance to β-lactam antibiotics
Class A β-Lactamase
Extended-spectrum β-lactamase (ESBLS) are the enzymes belongs to the Amber class A that confer resistant to most of the antibiotics such as penicillin, carbapenems and cephalosporin. The first report for isolation of ESBL producing strain was from Bangladesh in 200450. Apart from that different β-lactamases of the Amber class A such as TEM, SHV and CTX-M have been reported in Shigella spp. These enzymes are responsible for selective hydrolysis of ceftriaxone, cefotaxamine and ceftazidime51. Till now several reports from different parts of the world like Canada, Turkey, Israel, Argentina, China and India showed that Shigella spp. harbor different ESBL encoding genes (Table 1).
Class B β-Lactamase
Class B β-lactamase can hydrolyze carbapenem and other β-lactam. A study reported that, a metallo-β-lactamase (MET 1) encoded by a plasmid mediated gene IMP-3 in S. flexneri conferred resistance against sulfonamide and kanamycin. S. sonnei and S. flexneri isolated from Andaman and Nicobar Island in India showed the presence of blaVIM and blaIMP which conferred the resistance to carbapenem52.
Class C β-Lactamase
Ceftriaxone and cephalosporin has been recommended to treat the MDR Shigella spp. However Shigella spp. resistant to those antibiotics has also been reported. Class C β-Lactamase, known as AmpC-type enzymes are encoded by both chromosomal and plasmid genes. CMY-2, a plasmid encoded AmpC β-lactamase firstly identified in S. sonnei isolates which was obtained from a dysentery outbreak in Taiwan and there after CMY-2 have been reported in several countries like China, Iran, Costa Rica and India53-55. Different AmpC genes are listed in table1.
Class D β-Lactamase
Resistance to cloxacillin, ampicillin, cephalothin, oxacillin mainly mediated by class D β- lactamase or OXA-type β-lactamase56. OXA-type β-lactamase encoding gene blaOXA have been identified in integrons and plasmids in different Gram negative bacteria including Shigella spp. especially in S. flexneri57. blaOXA-1 and blaOXA-30 are differed from each other by having a single mutation at codon 131,containing Tn2603 and Tn1409 transposons respectively.
Table 1: Prevalence of antimicrobial resistance genes in Shigella spp. isolated from different regions of the world.
| Antimicrobial Class | Resistance Mechanism | Genes Mediating Antimicrobial Resistance | Origin | Geographic Origin | References |
| β-Lactams | Class A β-lactamases | blaSHV 2,11,12 | P,C | India, Argentina, | 80-82 |
| blaPER-2 | — | Argentina | 80 | ||
| blaTEM-1,1b,15,17
blaTEM-19,20,52 |
I, P | Lebanon, China, South Korea | 83-85 | ||
| blaCTX-M-1-123 | P,C | China, Turkey, Argentina, South Korea, India, Israel, Japan | 82,86-89 | ||
| Class B β-lactamases | blaIMP-like | P | India, France | 52 | |
| blaKPC | – | Senegal, France | 90 | ||
| blaVIM-like | – | India | 52 | ||
| Class C β-lactamases | blaCMY-2,59 | C, P | China, Mexico, India, | 53 | |
| blaDHA-1 | C, I, P | China, India, Israel | 86 | ||
| Class D β-lactamases | blaoxa-1,2,5,30 | I, P | Mozambique, Chile, China, India, US, Egypt, Djibouti, Spain, Greece, Denmark, Peru, Iran | 87,91 | |
| Quinolones | Plasmid-borne resistance | qnrA | P | Iran | 54 |
| qnrB,4,19 | P | India, Switzerland | 60,92 | ||
| qnrC | P | India | 93 | ||
| qnrS,I | P | Pakistan | 69 | ||
| aac-(60)-Ib-cr | P | India | 92 | ||
| Efflux pump | qepA | p | China | 60 | |
| Fosfomycin | Fosfomycin resistance enzymes | fosA3 | P | China | 64 |
| Aminoglycosides: streptomycin | strA,B | MGE | India, Australia, [1]Chile, Pakistan, South Korea | 67 | |
| Adenyltransferase | aadA1,2,5 | I, P | Senegal, Bhutan, India, Taiwan | 65,90,94 | |
| Tetracycline | Efflux pumps | tetA,B,G | C, P | Mozambique, Taiwan,Iran, Spain, South Korea | 9,94,95 |
| Trimethoprim | Dihydrofolate reductases | dfrA1,5,7,8,12,13
dfrA14,15,16,17 |
I, P | Spain, Taiwan, Senegal, Mozambique, India, China, Iran, South Korea, Peru, France, Chile, Australia | 9,90,91,94,7 |
| dfrV | I, P | China, India | |||
| Sulfonamides | Plasmid-borne resistance | sul1 | I, P | Australia, India | 65,67 |
| Phenicols | Chloramphenicol acetyltransferase genes | catA-like | P | Taiwan, Mozambique, India | 90 |
| catP | P | Pakistan | 69 | ||
| Efflux pumps | cmlA1 | I, P | Taiwan | 94 | |
| Colistin | Plasmid-borne resistance | mcr-1 | P | China, Vietnam | 73 |
| Macrolide | Enzymatic inactivation | mphA | P | Palestine, Switzerland | 60 |
| rRNA methylase | ermB | P | Vietnam | [2] |
Abbreviation: P, plasmid; C, chromosome; I, integrin; —, unknown; MGE, mobile genetic element.
Quinolones and fluroquinolones resistance
Resistance to fluroquinolones due to chromosomal target site mutations
Quinolones are used to treat shigellosis for a very long time across the world58. This group of antibiotics mainly consists of ciprofloxacin, norfloxacin, ofloxacin, levofloxacin, nalidixic acid etc. There are various genes present into the chromosomes as well as plasmids of the bacteria Shigella which confer resistance against these drugs. They can be mainly classified into plasmid mediated quinolone resistance genes (PMQR) and quinolone resistance determining regions (QRDR). Efflux pump attributor proteins also confer resistance to quinolones of the bacteria. gyrA, gyrB, parC and parE are the corresponding subunits of DNA gyrase and topoisomerase IV, which are encoded by gyrA, gyrB, parC and parE gene respectively (Table 1).Quinolones bind with QRDR region of DNA gyrase and mutation in that region causes reduced susceptibility to the quinolones. The most mutations have been found in between Ala 67 and Gln 07 in several studies59. A very recent study reported the mutations at the position 83 (Ser-83-Leu) and 87 (Asp-87-Asn) of gyrA in the Shigella spp. isolated from environmental water samples of in and around Kolkata have higher MIC to quinolones27 . Some researchers reported that a single mutation in gyrA cannot decrease the susceptibility to quinolone and for that further mutation in parC and gyrA region are needed60,61. Amino acid and nucleic acid changes in QRDR region of gyrA, gyrB, parC and parE in Shigella spp. are shown in table 2.
Table 2: Frequency of amino-acid and nucleotide changes in the quinolone resistance determining regions of Shigella isolates in different parts of the world.
| Target Site Mutations | Codon | Amino- Acid Changes | Nucleotide Mutation | Shigella spp. | Country Of Detection | References |
| gyrA | 57 | Asn→Lys | AAT→AAA | S. flexneri | China | 99 |
| 69 | Gln→Trp | — | S. sonnei | India | 93 | |
| 80 | His→ Gly | CAT→GGT | S. dysenteriae | Belgium | 99 | |
| 83 | Ser→Leu | TCG →TTG | S. boydii | China, Bangladesh, India | 92 | |
| 180 | Asp→Met | — | S165/15 | India | 27 | |
| 83 | Ser→Leu | — | S148/17 | India | ||
| 181 | Glu→Lys | — | S74/15 | India | ||
| 87 | Asp→Asn | — | S138/16 | India | ||
| 192 | Phe→His | — | S28/14 | India | ||
| 25 | Ile→Met | — | S124/16 | India | ||
| 39 | Glu→Lys | — | S49/15 | India | ||
| gyrB | 517 | Gln→Arg | CAG→CGA | S. flexneri | China | 61 |
| parC | 64 | Ala→Asp | GCC→GAC | S. flexneri, | India, China | 93 |
| 64 | Ala→Cys | GCC→TGC | S. sonnei | China | 93 | |
| 80 | Ser→Ile | AGC→ATC | S. dysenteriae | China, Bangladesh, India | 92 | |
| 85 | Ala→Ser | GCG→TCG | S. boydii | Switzerland | 60 | |
| parE | 408 | Gly→Asp | GGC→GAC | S. dysenteriae | China | 61 |
| 458 | Ser→Ala | TCG→GCG | S. flexneri | India | 61[1] |
[1] Note: —, unknown.
At parE two novel mutations at codons 408 and 458 have recently been identified among the Shigella spp. isolated in India (2011) and in China (2016)28,62. Mutation in codon 408 is associated with resistance to nalidixic acid but not to ciprofloxacin whereas mutation at codon 458 conferred resistance to both nalidixic acid and ciprofloxacin.
Plasmid mediated resistance
Presence of plasmid mediated gene namely qnr (qnrA, qnrB, qnrC, qnrD, qnrS, qep, aac[6’] –lb–cr) at the plasmid-mediated quinolone resistance region (PMQR) is also one of the main reason to gain resistance to the quinolones among the Shigella spp. A study from China reported that aac(6’)-lb-cr and qepA positive showed a high level of resistance to quinolones49. Gene aac (6’)-lb-cr encodes an acetyltransferase which reduce the activity of quinolones. Although the mutation in the QRDRs of DNA gyrase and topoisomerase IV genes is the main cause of resistance to fluroquinolones but PMQRs may expedite in selection of isolates showing higher levels of resistance through extra chromosomally encoded mechanisms and admit reduced susceptibility to fluroqinolones63 .
Fosfomycin resistance
Fosfomycin inhibit cell-wall biosynthesis by inactivating the MurA enzymes64. Although fosfomycin have been used to treat microbial infections for four decades but fosfomycin resistance have also been found in several enteropathogens including Shigella spp. Resistance to fosfomycin occurred primarily by two mechanisms: mutations in the uhpA/T and glpT genes which encode proteins responsible for two carrier dependent system associated with the uptake of fosfomycin ; fosfomycin modifying enzymes containing two kinase(FomA, FomB) and three metallo enzymes (FosX, FosA and FosB). For the first time fosfomycin modifying enzymes was reported from China among the isolated strain of S. flexneri from patient’ samples64 .
Aminoglycoside resistance
Aminoglycosides are being used to treat various bacterial infections for long time. They are associated with different kinds of mechanisms such as enzymatic inactivation, ribosomal modifications and active efflux pump. Out of these mechanisms, aminoglycoside modifying enzymes are the most common in clinical settings65,66. Aminoglycoside adenyltransferase (aadA gene cassettes) is very significant in Shigella spp. which confers resistance to streptomycin and spectinomycin66,67. Different types of aadA gene cassette have been identified in enterobacteriaceae but aadA1 and aadA2 has high prevalence among the isolated Shigella spp.66.Genes strA and strB which encode aminoglycoside phosphotransferase are also well dispersed among the plasmid (IncFII and pNV-Y394) of the isolated Shigella spp.65,68,69 . A study from India reported that 100% and 88% of S. dysenteriae and S. flexneri harbored strA genes which confer resistance to streptomycin70.
Tetracycline resistance
Roberts et al. reports five tetracycline-efflux genes-tet(A), tet(B), tet(C), tet(D) and tet(G) with one ribosomal protection protein encoded by tet(M) that have been identified among the Shigella spp. Various tet genes flanked by transposases were identified in a ~20.4 kb of genomic island encoding MDR genes71. Identical MDR cassette was firstly identified in S. flexneri 2a YSH6000 strain which is referred to as Shigella resistance locus-pathogenicity island71. Moreover presence of the MDR genes also reported in the E. coli plasmid pRs225 with similar arrangement which suggests that tet genes might be dispersed among the other species by horizontal gene transfer. S. dysenteriae isolated from dysentery outbreaks in different parts of India showed that tet(B) was more common (90%) than tet(A) among the isolates72.
Phenicol resistance
Phenicols have used for past few years to treat Shigella infection but the treatment is more challenging now a days due to resistance to these antibiotics. Resistance to chloramphenicol in Shigella spp. is associated with the cat genes [catA (catA1,catA2, catA3) and catB (catB2, catB3, catB7, catB8)] encoding chloramphenicol acetyltransferase, activation of efflux pump by cmlA (cmlA1, cmlA4, cmlA9) genes and/or by fluorinated and unfluorinated phenicols (flor) by major facilator-superfamily proteins73. About 96 Shigella were isolated from diarrheal patients sample in Pakistan and out of them 69 (72.9%) were resistant to chloramphenicol11
Colistin resistance
Colistin (polymixin E) interacts with the outer membranes of Gram negative bacteria. The gene responsible for colistin is a plasmid-mediated polymyxin resistance gene namely in mcr-1 have been identified in the Shigella spp.74. Gene mcr-1 produce a phosphatidyl ethanolamine which modify the lipid A on cell membranes and reduce the affinity for colistin and related polymixins and consequently reduce the antibacterial activity of these drugs75. Isolated Shigella spp. having mcr-1 gene showed four to eight fold increase in the MIC of polymixin B76. mcr-1 have been identified in S. sonnei isolated from Shanghai (2010-2012) which were resistant to polymixin B (MIC 4-8 µg/ml)76.
Sulfonamide and trimethoprim resistance
Spreading of trimethoprim-sulfonamide resistance among the Shigella spp. across the different parts of the world make this drug ineffective to treat shigellosis30. This is mainly due to mutational or recombinational changes in target enzymes (dihydropteroate synthase and dihydrofolate reductase respectively). The genes responsible for encoding dihydropteroate synthase and dihydrofolate reductase are sul and dfr respectively. Almost 42 types of dfr genes have been detected among different groups of bacteria which confers resistance to trimethoprim and 12 of them have been identified among the trimethoprim resistant Shigella spp. Gene cassettes within class 1 integrons among the Shigella plasmid or chromosome often encodes resistance to trimethoprim (dfrA), streptinomycin (aadA) and ampicillin (oxa-1)77. Class 2 integrons borne on Tn7 have often been found in Shigella spp., and gene cassette arrays of them usually contain dfrA1, sat1 and aadA1. Resistance to trimethoprim is mainly associated with the presence of dfrA1 genes occurring in a cassette in both class 1 and class 2 integrons. This integron associated antibiotic resistance may transfer to other species via plasmid conjugation. Gene cassette array carried by class 1 integron have been recognized in S. sonnei isolated from Chaina, Vietnam and Australia66,77. The genes responsible for sulfonamide resistance are sul1, sul2 and sul3, very common in Shigella spp. Different studies from the different parts of the world showed that number of Shigella spp. especially S. sonnei isolated from 2000 onwards have 100% resistance to the sulfonamides.
Macrolide resistance
Currently WHO recommends azithromycin as a second line treatment for shigellosis but now a days this antibiotics are becoming resistant day by day. CDC reports that approximately 3% of the all tested Shigella spp. is getting resistant to azithromycin. Possibly there are four mechanisms which mediate the resistance to these drugs including enzymatic inactivation by phosphotransferase encoded by mph gene or esterase encoded by ere determinant; target site modification by rRNA methylase encoded by erm genes; punctual mutation in rpIV encoding L22 ribosomal protein, rplD encoding L4 ribosomal protein and rrlH (23 rRNA); drug resistance mediated by efflux pumps including OmpA, OmpW, mefA and msrA78. Reduced susceptibility to azithromycin among the isolated Shigella spp. have been continuously reported from different parts of the world such as Asia, North-America, Australia and US79.
Thus frequent shifts in antimicrobial resistance profiles of Shigella isolates caused difficulty in recommending standard drugs for effective treatment of the disease.
Conclusion
Shigellosis is one of the most important health concerns in countries that yet to overcome many socioeconomic challenges. There are several virulence factors and enterotoxins which confer the pathogenicity of the bacteria Shigella, the causative agent of this disease. There were several antibiotics like ampicillin, tetracycline, foscomycin, trimethoprim, sulfonamide, macrolide group which now became ineffective. Only some fluoroquinolone group of antibiotics is active to combat the disease. But indiscriminate use of antibiotics and other risk factors help to develop a number of antibiotic resistant strains of Shigella spp. into the environment. This difficult situation demands the discovery of new and better drugs. So, it is very much essential to search for the actual mechanisms of developing drug resistance into the bacterial cells. In this review, we summarized the recent advancement of knowledge regarding different drugs used to combat the disease, latest pattern of drug resistance, development of antibiotic resistance mechanisms of Shigella spp. for better understanding of the host microbe interaction and to build up new strategy to combat this disease.
Acknowledgment
Authors thank to DST-SERB, Govt. of India and Department of Biotechnology West Bengal Govt. for financial assistance.
Conflicts of interest
All authors declare that they have no conflict of interest.
Funding Sources
The work was supported by DST-SERB, Govt. of India.
References
- Bhattacharya D, Bhattacharya H, Sayi DS, Bharadwaj AP, Singhania M, Sugunan AP, et al. Changing patterns and widening of antibiotic resistance in Shigella spp. over a decade (2000-2011), Andaman Islands, India. Epidemiology and infection 2015; 143 (3):470-477.
CrossRef - Karen L Kotloff, Mark S Riddle, James A Platts-Mills, Patricia Pavlinac, Anita K M Zaidi. Shigellosis. The Lancet 2017; 6736 (17):33296-8.
- Lamberti LM, Bourgeois AL, Fischer Walker CL, Black RE, Sack D. Estimating diarrheal illness and deaths attributable to Shigellae and enterotoxigenic Escherichia coli among older children, adolescents, and adults in South Asia and Africa. PLoS neglected tropical diseases 2014; 8 (2):e2705.
CrossRef - Shen H, Chen J, Xu Y, Lai Z, Zhang J, Yang H, et al. An outbreak of shigellosis in a Children Welfare Institute caused by a multiple-antibiotic-resistant strain of Shigella flexneri 2a. Journal of infection and public health 2017; 10 (6):814-818.
CrossRef - Baird-Parker AC. 1993 Fred Griffith Review Lecture. Foods and microbiological risks. Microbiology 1994; 140 ( Pt 4):687-695.
CrossRef - Zhang WX, Chen HY, Tu LH, Xi MF, Chen M, Zhang J. Fluoroquinolone Resistance Mechanisms in Shigella Isolates in Shanghai, China, Between 2010 and 2015. Microbial drug resistance 2019; 25 (2):212-218.
CrossRef - Puzari M, Sharma M, Chetia P. Emergence of antibiotic resistant Shigella species: A matter of concern. Journal of infection and public health 2017.
CrossRef - Taneja N, Mewara A. Shigellosis: Epidemiology in India. The Indian journal of medical research 2016; 143 (5):565-576.
CrossRef - Shahsavan S, Owlia P, Rastegar Lari A, Bakhshi B, Nobakht M. Investigation of Efflux-Mediated Tetracycline Resistance in Shigella Isolates Using the Inhibitor and Real Time Polymerase Chain Reaction Method. Iranian journal of pathology 2017; 12 (1):53-61.
CrossRef - GS WHO. Diarrhoea: Why children are still dying and what can be done. UNICEF/WHO 2009.
- Tariq A, Haque A, Ali A, Bashir S, Habeeb MA, Salman M, et al. Molecular profiling of antimicrobial resistance and integron association of multidrug-resistant clinical isolates of Shigella species from Faisalabad, Pakistan. Canadian journal of microbiology 2012; 58 (9):1047-1054.
CrossRef - GS WHO. Initiative for Vaccines Research, Diarrhoeal Diseases WHO tech- nical report series 2009.
- Bardhan P, Faruque AS, Naheed A, Sack DA. Decrease in shigellosis-related deaths without Shigella spp.-specific interventions, Asia. Emerging infectious diseases 2010; 16 (11):1718-1723.
CrossRef - Yabuuchi E. Bacillus dysentericus (sic) 1897 was the first taxonomic rather than Bacillus dysenteriae 1898. International journal of systematic and evolutionary microbiology 2002; 52 (Pt 3):1041.
CrossRef - wilkins Wa. Bergeys manual of determinative bacteriology. 1930; 3rd edition:357-363.
- Wei J, Goldberg MB, Burland V, Venkatesan MM, Deng W, Fournier G, et al. Complete genome sequence and comparative genomics of Shigella flexneri serotype 2a strain 2457T. Infection and immunity 2003; 71 (5):2775-2786.
CrossRef - Ostroi P, Anis E, Green MS. Shigellosis in Israel–update 1995. Public health reviews 1996; 24 (3-4):213-225.
- Taneja N, Lyngdoh V, Vermani A, Mohan B, Rao P, Singh M, et al. Re-emergence of multi-drug resistant Shigella dysenteriae with added resistance to ciprofloxacin in north India & their plasmid profiles. The Indian journal of medical research 2005; 122 (4):348-354.
- Muthuirulandi Sethuvel DP, Devanga Ragupathi NK, Anandan S, Veeraraghavan B. Update on: Shigella new serogroups/serotypes and their antimicrobial resistance. Letters in applied microbiology 2017; 64 (1):8-18.
CrossRef - El-Gendy A, El-Ghorab N, Lane EM, Elyazeed RA, Carlin NI, Mitry MM, et al. Identification of Shigella flexneri subserotype 1c in rural Egypt. Journal of clinical microbiology 1999; 37 (3):873-874.
CrossRef - Talukder KA, Islam MA, Dutta DK, Hassan F, Safa A, Nair GB, et al. Phenotypic and genotypic characterization of serologically atypical strains of Shigella flexneri type 4 isolated in Dhaka, Bangladesh. Journal of clinical microbiology 2002; 40 (7):2490-2497.
CrossRef - Kumar A OA, Alexander VS. Prevalence and antimicrobial susceptibility patterns of Shigella in stool samples in a tertiary healthcare hospital of Punjab. J Heal Res 2014; 1:33–35.
CrossRef - Reller ME, Nelson JM, Molbak K, Ackman DM, Schoonmaker-Bopp DJ, Root TP, et al. A large, multiple-restaurant outbreak of infection with Shigella flexneri serotype 2a traced to tomatoes. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America 2006; 42 (2):163-169.
CrossRef - Jiang Y, Yang F, Zhang X, Yang J, Chen L, Yan Y, et al. The complete sequence and analysis of the large virulence plasmid pSS of Shigella sonnei. Plasmid 2005; 54 (2):149-159.
CrossRef - Venkatesan MM, Ranallo RT. Live-attenuated Shigella vaccines. Expert review of vaccines 2006; 5 (5):669-686.
CrossRef - Ansaruzzaman M, Sultana M, Talukder KA, Alam K, Matsushita S, Safa A, et al. Isolation and characterization of provisional serovar Shigella boydii E16553 from diarrhoeal patients in Bangladesh. Journal of medical microbiology 2005; 54 (Pt 5):477-480.
CrossRef - Roy B, Tousif Ahamed SK, Bandyopadhyay B, Giri N. Development of quinolone resistance and prevalence of different virulence genes among Shigella flexneri and Shigella dysenteriae in environmental water samples. Letters in applied microbiology 2019.
CrossRef - Bhattacharya D, Bhattacharya H, Thamizhmani R, Sayi DS, Reesu R, Anwesh M, et al. Shigellosis in Bay of Bengal Islands, India: clinical and seasonal patterns, surveillance of antibiotic susceptibility patterns, and molecular characterization of multidrug-resistant Shigella strains isolated during a 6-year period from 2006 to 2011. European journal of clinical microbiology & infectious diseases : official publication of the European Society of Clinical Microbiology 2014; 33 (2):157-170.
CrossRef - Rudnai O, Csorian E, Lanyi B, Adam MM, Milch H. Salmonella and shigella surveillance in Hungary 1972-1976. I. Salmonella surveillance. Acta microbiologica Academiae Scientiarum Hungaricae 1981; 28 (1):37-52.
CrossRef - Kotloff KL, Platts-Mills JA, Nasrin D, Roose A, Blackwelder WC, Levine MM. Global burden of diarrheal diseases among children in developing countries: Incidence, etiology, and insights from new molecular diagnostic techniques. Vaccine 2017; 35 (49 Pt A):6783-6789.
CrossRef - Niyogi SK. Shigellosis. Journal of microbiology 2005; 43 (2):133-143.
CrossRef - Gupta AK, Nayduch D, Verma P, Shah B, Ghate HV, Patole MS, et al. Phylogenetic characterization of bacteria in the gut of house flies (Musca domestica L.). FEMS microbiology ecology 2012; 79 (3):581-593.
- Saha T, Murhekar M, Hutin YJ, Ramamurthy T. An urban, water-borne outbreak of diarrhoea and shigellosis in a district town in eastern India. The National medical journal of India 2009; 22 (5):237-239.
- Debnath F, Mukhopadhyay AK, Chowdhury G, Saha RN, Dutta S. An Outbreak of Foodborne Infection Caused by Shigella sonnei in West Bengal, India. Japanese journal of infectious diseases 2018; 71 (2):162-166.
CrossRef - Ranjbar R, Hosseini MJ, Kaffashian AR, Farshad S. An outbreak of shigellosis due to Shigella flexneri serotype 3a in a prison in Iran. Archives of Iranian medicine 2010; 13 (5):413-416.
- Perdomo JJ, Gounon P, Sansonetti PJ. Polymorphonuclear leukocyte transmigration promotes invasion of colonic epithelial monolayer by Shigella flexneri. The Journal of clinical investigation 1994; 93 (2):633-643.
CrossRef - Le-Barillec K, Magalhaes JG, Corcuff E, Thuizat A, Sansonetti PJ, Phalipon A, et al. Roles for T and NK cells in the innate immune response to Shigella flexneri. Journal of immunology 2005; 175 (3):1735-1740.
CrossRef - Gall TL, Mavris M, Martino MC, Bernardini ML, Denamur E, Parsot C. Analysis of virulence plasmid gene expression defines three classes of effectors in the type III secretion system of Shigella flexneri. Microbiology 2005; 151 (Pt 3):951-962.
CrossRef - Nandy S, Mitra U, Rajendran K, Dutta P, Dutta S. Subtype prevalence, plasmid profiles and growing fluoroquinolone resistance in Shigella from Kolkata, India (2001-2007): a hospital-based study. Tropical medicine & international health : TM & IH 2010; 15 (12):1499-1507.
CrossRef - Raja SB, Murali MR, Devaraj SN. Differential expression of ompC and ompF in multidrug-resistant Shigella dysenteriae and Shigella flexneri by aqueous extract of Aegle marmelos, altering its susceptibility toward beta-lactam antibiotics. Diagnostic microbiology and infectious disease 2008; 61 (3):321-328.
CrossRef - Kim JY, Kim SH, Jeon SM, Park MS, Rhie HG, Lee BK. Resistance to fluoroquinolones by the combination of target site mutations and enhanced expression of genes for efflux pumps in Shigella flexneri and Shigella sonnei strains isolated in Korea. Clinical microbiology and infection : the official publication of the European Society of Clinical Microbiology and Infectious Diseases 2008; 14 (8):760-765.
CrossRef - Kar AK, Ghosh AS, Chauhan K, Ahamed J, Basu J, Chakrabarti P, et al. Involvement of a 43-kilodalton outer membrane protein in beta-lactam resistance of Shigella dysenteriae. Antimicrobial agents and chemotherapy 1997; 41 (10):2302-2304.
CrossRef - Tran ENH, Papadopoulos M, Morona R. Relationship between O-antigen chain length and resistance to colicin E2 in Shigella flexneri. Microbiology 2014; 160 (Pt 3):589-601.
CrossRef - Sun J, Deng Z, Yan A. Bacterial multidrug efflux pumps: mechanisms, physiology and pharmacological exploitations. Biochemical and biophysical research communications 2014; 453 (2):254-267.
CrossRef - Yang H, Duan G, Zhu J, Lv R, Xi Y, Zhang W, et al. The AcrAB-TolC pump is involved in multidrug resistance in clinical Shigella flexneri isolates. Microbial drug resistance 2008; 14 (4):245-249.
CrossRef - Rahman M, Shoma S, Rashid H, Siddique AK, Nair GB, Sack DA. Extended-spectrum beta-lactamase-mediated third-generation cephalosporin resistance in Shigella isolates in Bangladesh. The Journal of antimicrobial chemotherapy 2004; 54 (4):846-847.
CrossRef - Bialvaei AZ, Pourlak T, Aghamali M, Asgharzadeh M, Gholizadeh P, Kafil HS. The Prevalence of CTX-M-15 Extended-spectrum beta-Lactamases Among Salmonella spp. and Shigella spp. Isolated from three Iranian Hospitals. European journal of microbiology & immunology 2017; 7 (2):133-137.
CrossRef - Thamizhmani R, Rhagavan R, Sugunan AP, Vijayachari P. VIM- and IMP-type metallo-beta-lactamase-producing Shigella spp. in childhood diarrhea from Andaman Islands. Infectious diseases 2015; 47 (10):749-750.
CrossRef - Tajbakhsh M, Garcia Migura L, Rahbar M, Svendsen CA, Mohammadzadeh M, Zali MR, et al. Antimicrobial-resistant Shigella infections from Iran: an overlooked problem? The Journal of antimicrobial chemotherapy 2012; 67 (5):1128-1133.
CrossRef - Ayala AT, Acuna HM, Calvo MT, Morales JL, Chacon EC. [Emergence of CMY-2-type plasmid-mediated AmpC beta-lactamase in Shigella sonnei and Salmonella spp. in Costa Rica, 2003-2015]. Revista panamericana de salud publica = Pan American journal of public health 2016; 40 (1):70-75.
- Zamanlou S, Ahangarzadeh Rezaee M, Aghazadeh M, Ghotaslou R, Babaie F, Khalili Y. Characterization of integrons, extended-spectrum beta-lactamases, AmpC cephalosporinase, quinolone resistance, and molecular typing of Shigella spp. from Iran. Infectious diseases 2018; 50 (8):616-624.
CrossRef - Cui X, Wang J, Yang C, Liang B, Ma Q, Yi S, et al. Prevalence and antimicrobial resistance of Shigella flexneri serotype 2 variant in China. Frontiers in microbiology 2015; 6:435.
CrossRef - Cui X, Yang C, Wang J, Liang B, Yi S, Li H, et al. Antimicrobial Resistance of Shigella flexneri Serotype 1b Isolates in China. PloS one 2015; 10 (6):e0129009.
CrossRef - Mannion AJ, Martin HR, Shen Z, Buckley EM, Dzink-Fox JL, Garcia A, et al. Plasmid-Mediated Quinolone Resistance in Shigella flexneri Isolated From Macaques. Frontiers in microbiology 2018; 9:311.
CrossRef - Azmi IJ, Khajanchi BK, Akter F, Hasan TN, Shahnaij M, Akter M, et al. Fluoroquinolone resistance mechanisms of Shigella flexneri isolated in Bangladesh. PloS one 2014; 9 (7):e102533.
CrossRef - Tamanna, Ramana J. Structural Insights into the Fluoroquinolone Resistance Mechanism of Shigella flexneri DNA Gyrase and Topoisomerase IV. Microbial drug resistance 2016; 22 (5):404-411.
CrossRef - Nuesch-Inderbinen M, Heini N, Zurfluh K, Althaus D, Hachler H, Stephan R. Shigella Antimicrobial Drug Resistance Mechanisms, 2004-2014. Emerging infectious diseases 2016; 22 (6):1083-1085.
CrossRef - Xue C, Cai J, Kang H, Chen Y, Wang K, Qian H, et al. Two novel mutations in parE among Shigella flexneri isolated from Jiangsu Province of China, 2016. Annals of translational medicine 2018; 6 (15):306.
CrossRef - Zhu Z, Cao M, Zhou X, Li B, Zhang J. Epidemic characterization and molecular genotyping of Shigella flexneri isolated from calves with diarrhea in Northwest China. Antimicrobial resistance and infection control 2017; 6:92.
CrossRef - Liu Y, Cheng Y, Yang H, Hu L, Cheng J, Ye Y, et al. Characterization of Extended-Spectrum beta-Lactamase Genes of Shigella flexneri Isolates With Fosfomycin Resistance From Patients in China. Annals of laboratory medicine 2017; 37 (5):415-419.
CrossRef - Shaw PC, Liang AC, Kam KM, Ling JM. Presence of strA-strB gene within a streptomycin-resistance operon in a clinical isolate of Shigella flexneri. Pathology 1996; 28 (4):356-358.
CrossRef - McIver CJ, White PA, Jones LA, Karagiannis T, Harkness J, Marriott D, et al. Epidemic strains of Shigella sonnei biotype g carrying integrons. Journal of clinical microbiology 2002; 40 (4):1538-1540.
CrossRef - Michael GB, Schwarz S. Antimicrobial resistance in zoonotic nontyphoidal Salmonella: an alarming trend? Clinical microbiology and infection : the official publication of the European Society of Clinical Microbiology and Infectious Diseases 2016; 22 (12):968-974.
CrossRef - Muthuirulandi Sethuvel DP, Anandan S, Devanga Ragupathi NK, Gajendiran R, Kuroda M, Shibayama K, et al. IncFII plasmid carrying antimicrobial resistance genes in Shigella flexneri: Vehicle for dissemination. Journal of global antimicrobial resistance 2019; 16:215-219.
CrossRef - Parajuli P, Deimel LP, Verma NK. Genome Analysis of Shigella flexneri Serotype 3b Strain SFL1520 Reveals Significant Horizontal Gene Acquisitions Including a Multidrug Resistance Cassette. Genome biology and evolution 2019; 11 (3):776-785.
CrossRef - Pazhani GP, Niyogi SK, Singh AK, Sen B, Taneja N, Kundu M, et al. Molecular characterization of multidrug-resistant Shigella species isolated from epidemic and endemic cases of shigellosis in India. Journal of medical microbiology 2008; 57 (Pt 7):856-863.
CrossRef - Parajuli P, Rajput MI, Verma NK. Plasmids of Shigella flexneri serotype 1c strain Y394 provide advantages to bacteria in the host. BMC microbiology 2019; 19 (1):86.
CrossRef - Traa BS, Walker CL, Munos M, Black RE. Antibiotics for the treatment of dysentery in children. International journal of epidemiology 2010; 39 Suppl 1:i70-74.
CrossRef - Schwarz S, Kehrenberg C, Doublet B, Cloeckaert A. Molecular basis of bacterial resistance to chloramphenicol and florfenicol. FEMS microbiology reviews 2004; 28 (5):519-542.
CrossRef - Poirel L, Jayol A, Nordmann P. Polymyxins: Antibacterial Activity, Susceptibility Testing, and Resistance Mechanisms Encoded by Plasmids or Chromosomes. Clinical microbiology reviews 2017; 30 (2):557-596.
CrossRef - Hinchliffe P, Yang QE, Portal E, Young T, Li H, Tooke CL, et al. Insights into the Mechanistic Basis of Plasmid-Mediated Colistin Resistance from Crystal Structures of the Catalytic Domain of MCR-1. Scientific reports 2017; 7:39392.
CrossRef - Ma Q, Huang Y, Wang J, Xu X, Hawkey J, Yang C, et al. Multidrug-resistant Shigella sonnei carrying the plasmid-mediated mcr-1 gene in China. International journal of antimicrobial agents 2018; 52 (1):14-21.
CrossRef - Iversen J, Sandvang D, Srijan A, Cam PD, Dalsgaard A. Characterization of antimicrobial resistance, plasmids, and gene cassettes in Shigella spp. from patients in vietnam. Microbial drug resistance 2003; 9 Suppl 1:S17-24.
CrossRef - Zhang C, Zhang R, Yu Q, Chu X, Sun J, Liu Q. Decreased Susceptibility to Azithromycin Among Clinical Shigella Isolates from China. Microbial drug resistance 2017; 23 (5):596-601.
CrossRef - Ingle DJ, Easton M, Valcanis M, Seemann T, Kwong JC, Stephens N, et al. Co-circulation of Multidrug-resistant Shigella Among Men Who Have Sex With Men in Australia. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America 2019; 69 (9):1535-1544.
CrossRef - Andres P, Petroni A, Faccone D, Pasteran F, Melano R, Rapoport M, et al. Extended-spectrum beta-lactamases in Shigella flexneri from Argentina: first report of TOHO-1 outside Japan. International journal of antimicrobial agents 2005; 25 (6):501-507.
CrossRef - Camacho AI, Irache JM, de Souza J, Sanchez-Gomez S, Gamazo C. Nanoparticle-based vaccine for mucosal protection against Shigella flexneri in mice. Vaccine 2013; 31 (32):3288-3294.
CrossRef - Liu G, Qian H, Tang B, Chen Y, Kang H, Jiang F, et al. Prevalence and characterisation of third-generation cephalosporin-resistant Shigella flexneri isolates from Jiangsu Province, China, 2013-2015. Journal of global antimicrobial resistance 2018; 15:283-287.
CrossRef - Matar GM, Jaafar R, Sabra A, Hart CA, Corkill JE, Dbaibo GS, et al. First detection and sequence analysis of the bla-CTX-M-15 gene in Lebanese isolates of extended-spectrum-beta-lactamase-producing Shigella sonnei. Annals of tropical medicine and parasitology 2007; 101 (6):511-517.
CrossRef - Kim JS, Kim J, Jeon SE, Kim SJ, Kim NO, Hong S, et al. Complete nucleotide sequence of the IncI1 plasmid pSH4469 encoding CTX-M-15 extended-spectrum beta-lactamase in a clinical isolate of Shigella sonnei from an outbreak in the Republic of Korea. International journal of antimicrobial agents 2014; 44 (6):533-537.
CrossRef - Kim S, Kim J, Kang Y, Park Y, Lee B. Occurrence of extended-spectrum beta-lactamases in members of the genus Shigella in the Republic of Korea. Journal of clinical microbiology 2004; 42 (11):5264-5269.
CrossRef - Vasilev V, Japheth R, Yishai R, Andorn N, Valinsky L, Navon-Venezia S, et al. Extended-spectrum beta-lactamase-producing Shigella strains in Israel, 2000-2004. European journal of clinical microbiology & infectious diseases : official publication of the European Society of Clinical Microbiology 2007; 26 (3):189-194.
CrossRef - Zhang CL, Liu QZ, Wang J, Chu X, Shen LM, Guo YY. Epidemic and virulence characteristic of Shigella spp. with extended-spectrum cephalosporin resistance in Xiaoshan District, Hangzhou, China. BMC infectious diseases 2014; 14:260.
CrossRef - Nagano Y, Nagano N, Wachino J, Ishikawa K, Arakawa Y. Novel chimeric beta-lactamase CTX-M-64, a hybrid of CTX-M-15-like and CTX-M-14 beta-lactamases, found in a Shigella sonnei strain resistant to various oxyimino-cephalosporins, including ceftazidime. Antimicrobial agents and chemotherapy 2009; 53 (1):69-74.
CrossRef - Barel LA, Mulard LA. Classical and novel strategies to develop a Shigella glycoconjugate vaccine: from concept to efficacy in human. Human vaccines & immunotherapeutics 2019; 15 (6):1338-1356.
CrossRef - Sambe-Ba B, Seck A, Wane AA, Fall-Niang NK, Gassama-Sow A. [Sensitivity to antibiotics and genetic support to resistance of Shigella flexneri strains isolated in Dakar from 2001 to 2010]. Bulletin de la Societe de pathologie exotique 2013; 106 (2):89-94.
CrossRef - Allue-Guardia A, Koenig SSK, Quiros P, Muniesa M, Bono JL, Eppinger M. Closed Genome and Comparative Phylogenetic Analysis of the Clinical Multidrug Resistant Shigella sonnei Strain 866. Genome biology and evolution 2018; 10 (9):2241-2247.
CrossRef - Dutta S, Kawamura Y, Ezaki T, Nair GB, Iida K, Yoshida S. Alteration in the GyrA subunit of DNA gyrase and the ParC subunit of topoisomerase IV in Quinolone-resistant Shigella dysenteriae serotype 1 clinical isolates from Kolkata, India. Antimicrobial agents and chemotherapy 2005; 49 (4):1660-1661.
CrossRef - Gu B, Qin TT, Fan WT, Bi RR, Chen Y, Li Y, et al. Novel mutations in gyrA and parC among Shigella sonnei strains from Jiangsu Province of China, 2002-2011. International journal of infectious diseases : IJID : official publication of the International Society for Infectious Diseases 2017; 59:44-49.
CrossRef - Ranjbar R, Aleo A, Giammanco GM, Dionisi AM, Sadeghifard N, Mammina C. Genetic relatedness among isolates of Shigella sonnei carrying class 2 integrons in Tehran, Iran, 2002-2003. BMC infectious diseases 2007; 7:62.
CrossRef - Wibberg D, Szczepanowski R, Eikmeyer F, Puhler A, Schluter A. The IncF plasmid pRSB225 isolated from a municipal wastewater treatment plant’s on-site preflooder combining antibiotic resistance and putative virulence functions is highly related to virulence plasmids identified in pathogenic E. coli isolates. Plasmid 2013; 69 (2):127-137.
CrossRef - Bischoff KM, White DG, Hume ME, Poole TL, Nisbet DJ. The chloramphenicol resistance gene cmlA is disseminated on transferable plasmids that confer multiple-drug resistance in swine Escherichia coli. FEMS microbiology letters 2005; 243 (1):285-291.
CrossRef - Seol SY, Kim YT, Jeong YS, Oh JY, Kang HY, Moon DC, et al. Molecular characterization of antimicrobial resistance in Shigella sonnei isolates in Korea. Journal of medical microbiology 2006; 55 (Pt 7):871-877.
CrossRef - Iqbal MS, Rahman M, Islam R, Banik A, Amin MB, Akter F, et al. Plasmid-mediated sulfamethoxazole resistance encoded by the sul2 gene in the multidrug-resistant Shigella flexneri 2a isolated from patients with acute diarrhea in Dhaka, Bangladesh. PloS one 2014; 9 (1):e85338.
CrossRef - Qin T, Bi R, Fan W, Kang H, Ma P, Gu B. Novel mutations in quinolone resistance-determining regions of gyrA, gyrB, parC and parE in Shigella flexneri clinical isolates from eastern Chinese populations between 2001 and 2011. European journal of clinical microbiology & infectious diseases : official publication of the European Society of Clinical Microbiology 2016; 35 (12):2037-2045.
CrossRef

This work is licensed under a Creative Commons Attribution 4.0 International License.