Manuscript accepted on : 21-12-2021
Published online on: 23-12-2021
Plagiarism Check: Yes
Reviewed by: Dr. Alaa Kareem Niamah
Second Review by: Dr. Biswajit Batabyal
Final Approval by: Dr Jahwarhar Izuan Bin Abdul Rashid
Najwa Menwer Alharbi*, Amjad Khalid Alharthi , Alsamadani
, Alsamadani , Raneem Ahmed Almihmadi
, Raneem Ahmed Almihmadi and Bothaina Ali Alaidaroos
and Bothaina Ali Alaidaroos
King Abdelaziz University, Science College, Biology Department, Jeddah, Saudi Arabia
Corresponding Author E-mail: Nmaalharbi@kau.edu.sa
DOI : http://dx.doi.org/10.13005/bbra/2954
ABSTRACT:
This study aimed to investigate a method to manage antimicrobial resistance (AMR) issues by exploring soil microorganisms that are capable of producing bioactive compounds. Eight different types of soil were selected from three locations to screen, isolate, and identify microorganisms that are capable of producing antimicrobial compounds. The multi-drug resistant strains are Staphylococcus aureus, Pseudomonas aeruginosa, Escherichia coli, and Candida albicans were selected for primary and secondary screening using the crowded plate method and the agar well diffusion method. Inhibition zones were measured, and data were assessed using statistical tests to check for normality and differences between parametric variables and nonparametric data. This was followed by biochemical characterization, DNA isolation, and polymerase chain reaction (PCR). Molecular identification was performed using 16S rRNA amplification and sequencing. Here, 86 isolates containing bacteria and fungi were successfully extracted from soil samples. Further, 49 of 86 microbes showed possible antimicrobial activity, but only 12 isolates resulted in distinct inhibition zones with the selected multi-drug resistant strains. The following different taxa were identified: Firmicutes (nine strains), Proteobacteria (one strain), Actinobacteria (one strain), and Azotobacter (one strain). Species are represented in a phylogenetic tree, which was constructed using the unweighted pair-group method with arithmetic mean (UPGMA) method. The evolutionary distances were computed using the Maximum Composite Likelihood method. The identified microorganisms showed antimicrobial activity, confirming that soil microorganisms have great potential to address AMR issues.
KEYWORDS: Antibiotic; Antibiotic Resistance; Soil Microbes
Download this article as:| Copy the following to cite this article: Alharbi N. M, Alharthi A. K, Alsamadani A, Almihmadi R. A, Alaidaroos B. A. Screening and Characterization of Soil Microbes Producing Antimicrobial Compounds in Makkah Province, Saudi Arabia. Biosci Biotech Res Asia 2021;18(4). |
| Copy the following to cite this URL: Alharbi N. M, Alharthi A. K, Alsamadani A, Almihmadi R. A, Alaidaroos B. A. Screening and Characterization of Soil Microbes Producing Antimicrobial Compounds in Makkah Province, Saudi Arabia. Biosci Biotech Res Asia 2021;18(4). Available from: https://bit.ly/3su9bgU |
Introduction
The spread of antimicrobial resistance (AMR) is now a critical global issue that requires a serious response to combat its effect on human and animal health. Researchers worldwide are combining their efforts to discover and develop new vital antimicrobials to minimize this growing problem 1–3. Owing to the overuse of broad-spectrum antibiotics to treat infections in humans, their excessive use in agriculture, and poor infection control in hospitals, resistant pathogens have become a serious threat to public health 4–6 . As such, a database was procured to predict the existence of nearly more than 20,000 undiscovered resistance genes (r genes) of almost 400 different types of the r genes predicted in the main from available bacterial genome sequences 7, 8. Microorganisms such as Staphylococcus aureus, Candida albicans, Pseudomonas aeruginosa, and Escherichia coli are pathogens that are responsible for many life-threatening infections; hence finding a potent antimicrobial is urgently needed to hinder their spread 8–10.
According to the World Health Organization, by 2050, 10 million human lives will be lost annually, and the economy will be substantially affected due to this resistance issue 1, 10–12. Therefore, more serious precautions need to be taken to prevent this disaster. The 2015 National Coordinate Committee was implemented by the 68 World Health Assembly to create a global antimicrobial resistance plan that aims to arrest the impact of these pathogens. Consequently, the One Health Approach is used as part of the worldwide strategy endorsed in 2016 at the United Nations General assembly after discussing antimicrobial resistance 13.
Soil is considered a vital and diverse ecosystem for many microorganisms, including bacteria, with potential antimicrobial activity. Approximately 60% of the discovered antibiotics each year are obtained from the soil; hence soil microorganisms represent a promising source 14. Many microorganisms produce antimicrobial substances to protect themselves from the antagonistic behavior of other microbes in their environment. Saudi Arabia has many different soil types owing to its various terrains encompassing undiscovered habitats, which might provide possible novel antimicrobial-producing microbes; therefore, three different sites were selected as a source of soil samples. This study aimed to investigate a method to manage antimicrobial resistance (AMR) issues by exploring soil microorganisms that are capable of producing antimicrobial compounds.
Material and methods
Soil sampling
Soil samples (10–15 g) were collected from the topsoil layer (10–15 cm) in sterile labeled plastic bags using a sterile spatula after removing all surface debris. The bags were transported to the laboratory in an icebox and stored at 4 ºC until further analysis. The soil samples were collected from three different locations in Makkah province, Saudi Arabia. Two different samples were collected from Salam Alzawahir, a small village located on the west coast of Saudi Arabia that lies approximately 400 km south of Jeddah city, characterized by its harsh environment and high atmospheric temperature most of the year. One soil sample was collected from Salam Alzawahir mangrove swamps (19°47’47.6″N 40°43’50.0″E), as shown in Figure 1 (A), and the second was from a private farm (19°48’48.5″N 40°43’58.0″E). Five soil samples were collected from different locations in Barzah, a village located in the northeast of Jeddah, Saudi Arabia. Barzah is a part of Harrat Rahat, which is the largest volcanic field within Saudi Arabia, at 50–75 km wide and ∼300 km long 15. This area is an active volcanic field characterized by two historical eruptions that occurred at approximately 641 and 1256 AD 16. The first soil sample was collected from a local farm (21°57’19.398″N 39°41’33.678″E), as shown in Figure 1(B). The second sample was taken from red soil (21°56’59.67″N 39°43’9.858″E), depicted in Figure 1(C), and the third was from granite soil (21°57’3.63″N 39°43’3.348″E), shown in Figure 1(D). The fourth sample was taken from a low-level rainwater brook filled with tadpoles, from which mud was collected in a sterile container (21°54’54.09″N 39°53’26.298″E), outlined in Figure 1(E). The fifth soil sample was collected from an oasis (21°53’49.692″N 39°44’55.08″E), shown in Figure 1(F). Another soil sample was obtained from the sediments of Lake 40th, which receives Jeddah city wastewater discharge (Figure 1(G)).
Isolation, primary screening, purification, and maintenance of antimicrobial-producing soil microorganisms
Antimicrobial-producing soil bacteria were isolated using the well-known “Crowded plate” approach following a typical serial dilution. To obtain a 1:10 dilution, 1 g of each soil sample was weighed and soaked in 10 ml of sterile distilled water and then allowed to settle after vigorous shaking. After collecting the supernatant, it was serially diluted. Next, 100 µl of each dilution was aseptically spread onto the surface of labeled nutrient agar plates, and Actinomycete Agar, for bacterial isolation, or Sabouraud dextrose agar (SDA) and potato dextrose agar (PDA), for fungus isolation, 14 were inoculated using a sterile glass rod. Nutrient agar plates were incubated at 37 °C for 24 to 48 h, whereas SDA and PDA agar plates were incubated at 28 °C for 3–5 days. Distinct bacteria and Actinomycetes colonies with distinct margins were picked and isolated on TSA plates using a sterile disposable cotton swab and incubated at 37 °C for 24 h. Fungi with distinct margins were selected for isolation on SDA, and PDA was incubated at 25 °C for 4–5 days 14. Pure cultures were maintained at 4 °C for subsequent studies.
Morphological and biochemical characterization of bacterial isolates
Pure isolates that exhibited inhibition zones were subjected to Gram staining. After a 24 h incubation at 37 °C, the morphology of each bacterial colony on agar plates was evaluated, and individual colonies were classified based on color, shape, appearance, colony diameter, and transparency. To discriminate between gram-positive and gram-negative bacteria, Gram staining (Thermo Fisher Scientific, Massachusetts, USA) was utilized 17.
Biochemical screening of the isolated bacteria
The bacterial isolates were characterized biochemically to evaluate their chemical nature. We conducted oxidase, catalase, Voges-Proskauer, methyl red, indole production, starch hydrolysis, citrate utilization, and carbohydrate fermentation tests, in addition to assessing growth on MacConkey agar, according to the standard protocols.
Secondary screening for isolates with antimicrobial activity
The antimicrobial activity of the isolated strains was evaluated against four multi-drug resistance strains, including S. aureus, P. aeruginosa, E.coli, and C. albicans, obtained from King Abdulaziz University Hospital. Secondary screening for antimicrobial production ability of the purified isolates was performed using the agar well diffusion method with Mueller Hinton agar media. In brief, sterilized cotton buds were used to pick pathogen colonies and swabbed on MHA plates. A hole of 6–8 mm in diameter was punched on MHA four-well plates using a sterile cork borer. Isolated and sub cultured, unknown colonies were inoculated in test tubes containing 5 ml of nutrient broth, incubated at 37 °C for 24 h. Next, 100 µl of the culture supernatant was poured into the wells using a micropipette and kept in an incubator for 24–48 h; sterile nutrient broth was used in the negative control well 14. The diameter of each inhibited zone was measured, and the level of antagonism against test pathogens was determined. The experiment was conducted in triplicate (Figure 2). For each replicate, measurements in millimeters were obtained for statistical analysis.
Genomic DNA isolation and PCR amplification
Genomic DNA was extracted using the QIAamp-DNA-Mini-kit for Bacterial genomic DNA isolation (QIAGEN, Germany) according to the manufacturer’s instructions. DNA quality and quantity were evaluated using a Nanodrop Thermo Scientific™. The bacterial broth was centrifuged to discard the supernatant and obtain the cell pellet. The pellet was washed with 0.9% saline and suspended in the digestion buffer. The genomic DNA extraction process was performed using an automated DNA extractor (Invent Technologies Ltd., Dhaka, Bangladesh) according to the manufacturer’s instructions. The concentration of the isolated DNA was measured using a spectrophotometer at 260 and 280 nm wavelengths. The purity of genomic DNA was checked before further use.
16S rRNA amplification and sequencing for molecular identification of antimicrobial-producing bacteria
GoTaq® Green Master Mix (Promega Corporation, Wisconsin, USA) was employed to amplify the 16S rRNA gene fragments, as per the manufacturer’s instructions. Forward (5′-AGA GTT TGA TCM TGG CTC AG-3′) and reverse (5′-CGG TTA CCT TGT TAC GAC TT-3′) universal primers were employed. In summary, the PCR (25 µl reaction mixture) began with an initial denaturation of 95 °C for 5 min followed by 35 cycles with steps of 94 °C for 30 s, 55 °C for 45 s a final 5 min extension step at 72 °C. The amplified PCR products were run on a 1% agarose gel and documented using GoTaq® Green Master Mix (Promega Corporation, Wisconsin, USA) and ethidium bromide staining. Purified 16S rDNA fragments were sent for sequencing to King Fahd Research Center in Jeddah, Saudi Arabia. The obtained sequences were edited using BioEdit 7.2 and aligned using the NCBI BLAST service to identify matches against the NCBI GeneBank database in a range of 98–100% similarity.
Statistical analysis
Data were collected, organized, and presented in tables and figures using Microsoft Excel version 2016. Variables were checked for normality using the Shapiro-Wilk normality test at the 0.05 level. Accordingly, the differences between isolates based on nonparametric data were evaluated using a Kruskal-Wallis test; however, differences in parametric variables between studied species and isolates were assessed by one- and two-way analysis of variance at a 0.05 level. Duncan’s multiple range test was used after ANOVA to further compare isolates. Data analysis was carried out using IBM-SPSS version 26.0 for Mac OS.
Phylogenetic tree
Visual representation of phylogeny among identified microorganisms, as shown in Figure 3, was performed using MEGA X version 10.0 software for Windows 10 64-bit; sequences were aligned, and a phylogenetic tree was constructed.
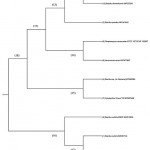 |
Figure 3: Phylogenetic tree structure represents the identified isolates’s species. |
Results
Screening, isolation, characterization, and molecular identification of soil antimicrobial-producing microorganisms
Soil samples were collected from different sites in Saudi Arabia (Salam Alzawahir village, Barzah village, and Lake 40th in Jeddah). The primary screening for antimicrobial-producing microorganisms was performed by the common crowded plate method, and presumptive antimicrobial-producing colonies were selected. Using the crowded plate method, 86 presumptive antimicrobial-producing colonies with a distinct morphology were selected and subcultured. Subsequently, the antimicrobial activities of the isolated strains were subjected to secondary screening against four different pathogens (S. aureus, C. albicans, P. aeruginosa, and E. coli) using the agar well diffusion procedure. Of 86 isolates, 12 bacterial strains were selected during the secondary screening process after showing distinct inhibition zones, and these were subjected to biochemical characterization and Gram staining (Table 1).
Molecular identification of the isolated strains through 16S rRNA gene sequencing was performed. The 12 isolates were classified as follows: Firmicutes (nine strains), Proteobacteria (one strain), Actinobacteria (one strain), and Azotobacter (one strain). According to the NCBI and BLAST gene bank, the identified bacterial strains were Bacillus subtilis (two isolates), B. licheniformis, Cytobacillus sp., Streptomyces sp., Aeromonas veronii, B. subtilis EBV2, Bacillus sp. ESA (two isolates), Bacillus strain 114, B. pumilus, and Bacillus sp HD1T) with 98–100% sequence similarity. As shown in Table 1, there were highly significant (p<0.001***) differences among isolates with respect to Gram stain and biochemical characteristics (including catalase and oxidase), as determined by the Kruskal-Wallis test. The identified microorganisms were B. subtilis, B. licheniformis, Cytobacillus sp., Streptomyces venezuelae, A. veronii, B. subtilis EBV2, Bacillus sp. ESA (two isolates), Bacillus strain 114, B. pumilus, and Bacillus sp HD1T (Table 1).
Table 1: Identification of bacterial isolates from various habitats. Differences between isolates were assessed via the Kruskal-Wallis test at P<0.05
| Isolate codes | Colonial morphology | Gram
stain |
Biochemical characteristic | Shape/ microscope | Media | Identified microorganism | ||||
| Whole colony | Surface texture | Edge | Pigment | Catalase | Oxidase | |||||
| R210 | Round | Smooth, sticky | Opaque | Creamy | +ve | +ve | +ve | Bacillus | TSA | Bacillus subtilis |
| Aj 11 | Irregular | Smooth | Umbonate | Translucent | +ve | +ve | +ve | Bacillus | SDA/PDA | Bacillus licheniformis |
| Aj1 | Round | Smooth | Entire | Creamy | -ve | -ve | +ve | Bacillus | SDA/PDA | Cytobacillus |
| Aj2 | Round | Wrinkled | Wet | Creamy | +ve | +ve | +ve | Bacillus | SDA/PDA | Streptomyces venezuelae |
| Aj 22 | Irregular | Smooth | Irregular | Dark green | -ve | +ve | +ve | Bacillus | SDA/PDA | Aeromonas veronii |
| Aj5 | Round | Wet | Wet | White | +ve | +ve | +ve | Bacillus | SDA/PDA | Bacillus subtilis |
| A41 | Cotton appearance | Irregular | White | +ve | +ve | -ve | Streptobacilli | Actinomycetes agar | Bacillus subtilis EBV2 | |
| A34 | Irregular | Smooth | Irregular | Creamy | +ve | +ve | -ve | Bacillus | Actinomycetes agar | Bacillus sp. ESA |
| A39 | Wet, rhizoid | Wet | Entire | Creamy | +ve | +ve | -ve | Streptobacilli | Actinomycetes agar | Bacillus sp. ESA |
| A24 | Irregular | Wrinkled | Irregular | Brownish | -ve | -ve | -ve | Coccus | PDA/NA | Bacterum strain 114 |
| A7 | Irregular | Wet | Cotton-like elevation | White | +ve | +ve | -ve | Bacillus | NA | Bacillus Pumilus |
| A8 | Filiform | Smooth | Filiform | Creamy | +ve | +ve | -ve | Bacillus | NA | Bacillus sp HD1T |
|
Descriptive statistics of various nonparametric measures |
Mean | 0.833 | 0.916 | 0.5 | ||||||
|
Median |
1 | 1 |
0.5 |
|||||||
| IQR | 1.0–1.0 | 1.0–1.0 |
0.5–1.0 |
|||||||
| Mode | 1 | 1 | 0.5 | |||||||
|
Kruskal-Wallis test |
P-value | <0.001*** | <0.001*** | <0.001*** | ||||||
*, **, *** Significant at p<0.05, <0.01, <0.001; non-significant at p>0.05
Note: +, 90% or greater positive; -, 90% or greater negative.
Biochemical characterization
The biochemical characteristics determined by the previous tests are listed in Table 1, which showed highly significant (p<0.001***) differences among tested isolates, as revealed by a Kruskal-Wallis test.
Screening for antimicrobial activity
Inhibition zones were observed. Of 86 pure isolates, 12 had antibiotic activity and were selected for further tests.
Microorganism antimicrobial activity determination
Table 2 presents 12 isolates coded against methicillin-resistant S. aureus, C. albicans, E. coli, and P. aeruginosa. Most isolates resulted in highly significant inhibition zones in the presence of methicillin-resistant S. aureus, which was followed by activity against C. albicans and E.coli, where no inhibition of P. aeruginosa was noted (Table 2). Against methicillin-resistant S. aureus, isolates showed a median (IQR) of 1 (1.0–1.0), and against C. albicans, a median (IQR) of 1.0 (0.0–1.0) was also observed (Table, 2).
Table 2: Isolates that exhibited inhibition zones
| No. | Isolates codes | Methicillin-resistance S. aureus | C. albicans | E. coli | P. aeruginosa |
| 1 | R210 | + | – | – | – |
| 2 | Aj11 | + | + | + | – |
| 3 | Aj1 | + | + | – | – |
| 4 | Aj2 | + | + | – | – |
| 5 | Aj22 | – | + | – | – |
| 6 | Aj5 | + | – | – | – |
| 7 | A41 | + | – | – | – |
| 8 | A34 | + | – | – | – |
| 9 | A39 | + | – | – | – |
| 10 | A24 | + | – | – | – |
| 11 | A7 | + | – | – | – |
| 12 | A8 | + | – | – | – |
| Descriptive | Mean | 0.91 | 0.34 | 0.09 | 0.00 |
| Median | 1.00 | 1.00 | 0.00 | 0.00 | |
| IQR | 1.0–1.0 | 0.0–1.0 | 0.0–0.0 | 0.0–0.0 | |
| Mode | 1.00 | 0.00 | 0.00 | 0.00 | |
| Kruskal-Wallis (p-value) | <0.001*** | <0.001*** | <0.001*** | >0.050 ns | |
*, **, *** Significant at p<0.05, <0.01, <0.001; ns, non-significant at p>0.05
Table 3: Staphylococcus aureus inhibition zones measured in triplicate for confirmation
| Isolates | Inhibition zone (mm) | ||
| Mean | SD | SE | |
| R210 | 20.7 | 8.5 | 4.91 |
| A7 | 22.0 | 4.6 | 2.65 |
| A8 | 19.7 | 5.1 | 2.96 |
| A24 | 21.7 | 2.1 | 1.20 |
| A39 | 19.3 | 4.6 | 2.67 |
| A41 | 15.7 | 7.2 | 4.18 |
| Aj11 | 21.0 | 1.0 | 0.58 |
| Aj2 | 18.7 | 1.5 | 0.88 |
| Aj1 | 18.0 | 2.0 | 1.15 |
| Aj5 | 19.0 | 2.0 | 1.15 |
| ANOVA | |||
| F-ratio | 0.518 | ||
| p-value | >0.05 ns | ||
*, **, *** Significant at p<0.05, <0.01, <0.001; ns, non-significant at p>0.05
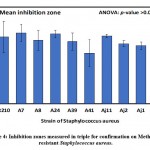 |
Figure 4: Inhibition zones measured in triple for confirmation on Methicillin-resistant Staphylococcus aureus. |
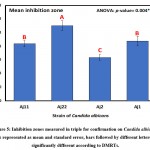
The results of clearing zones of S. aureus with the 10 isolates are presented in Table 3 and Figure 4, wherein the highest average (±SD) of 22.0±4.6 mm was found for the A7 isolate, followed by 21.7±2.1 mm for A24, and the smallest clearing zone of 15.7±7.2 mm for A41. Differences in inhibition zones between identified isolates were not significant (p>0.05) as revealed by one-way analysis of variance (Table, 3; Figure, 4). Furthermore, results of inhibition zones of C. albicans with the four tested isolates Aj22, Aj1, Aj11, and Aj2 showed averages (±SD) of 27.7±3.2, 22.0±3.0, 21.0±2.0, and 16.0±2.0 mm, respectively. A highly significant (p=0.004) difference in clearing zones among the four isolated was revealed by ANOVA. Aj22 showed the largest clearing zone, as revealed by Duncan’s multiple range tests (DMRTs), with significance at a 0.05 level (Table 4; Figure 5).
Table 4: Candida albicans inhibition zones (mm) measured in triplicate for confirmation
| Isolate | Mean | SD | SE |
| Aj11 | 21.0 b | 2.0 | 1.15 |
| Aj22 | 27.7 a | 3.2 | 1.86 |
| Aj2 | 16.0 c | 2.0 | 1.15 |
| Aj1 | 22.0 b | 3.0 | 1.73 |
| ANOVA | |||
| F-ratio | 10.05 | ||
| p-value | 0.004** | ||
*, **, *** Significant at p<0.05, <0.01, <0.001; non-significant at p>0.05
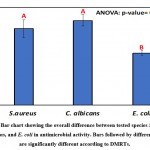
Comparatively, the overall average of inhibition zones of S. aureus, C. albicans, and E. coli with the isolates are presented in Table 5 and Figure 6. The largest clearing zones of 21.7±2.6 and 19.6±3.9 were observed for C. albicans and S. aureus as revealed by ANOVA and DMRTs. Table 6 and Figure 7 show the overall effects of isolates on S. aureus, C. albicans, and E. coli, and differences were assessed via two-way analysis of variance.
Table 5: Staphylococcus aureus, Candida albicans, and Escherichia coli inhibition zones (mm) with isolates of different species
| Species | Inhibition zone (mm) | ||
| Mean | SD | SE | |
| S. aureus | 19.6 a | 3.9 | 2.2 |
| C. albicans | 21.7 a | 2.6 | 1.5 |
| E. coli | 13.3 b | 1.0 | 0.6 |
| One-way ANOVA | |||
| F-ratio | 4.56 | ||
| p-value | 0.016* | ||
*, **, *** Significant at p<0.05, <0.01, <0.001; non-significant at p>0.05
Table 6: Overall differences among tested isolates for Staphylococcus aureus, Candida albicans, and Escherichia coli antimicrobial activity. Means followed by different letters are significantly different according to DMRTs.
| Species | Strains | Inhibition zone (mm) | ANOVA | ||||
| Mean | DMRTs | SD | SE | F-ratio | p-value | ||
| Staphylococcus aureus | R210 | 20.67 | abc | 8.50 | 4.91 | 0.518 | >0.05 ns |
| A7 | 22.00 | ab | 4.58 | 2.65 | |||
| A8 | 19.67 | bc | 5.13 | 2.96 | |||
| A24 | 21.67 | ab | 2.08 | 1.20 | |||
| A39 | 19.33 | bc | 4.62 | 2.67 | |||
| A41 | 15.67 | bc | 7.23 | 4.18 | |||
| Aj11 | 21.00 | abc | 1.00 | 0.58 | |||
| Aj2 | 18.67 | bc | 1.53 | 0.88 | |||
| Aj1 | 18.00 | bc | 2.00 | 1.15 | |||
| Aj5 | 19.00 | bc | 2.00 | 1.15 | |||
| Candida albicans | Aj11 | 21.00 | abc | 2.00 | 1.15 | 10.05 | 0.004** |
| Aj22 | 27.67 | a | 3.21 | 1.86 | |||
| Aj2 | 16.00 | bc | 2.00 | 1.15 | |||
| Aj1 | 22.00 | ab | 3.00 | 1.73 | |||
|
Escherichia coli |
— | 13.33 | c | 1.00 | 0.58 | — | — |
| Two-way ANOVA | |||||||
| Corrected model | 0.040 * | ||||||
| Intercept | <0.001*** | ||||||
| Isolates | 0.040 * | ||||||
| Species | 0.016* | ||||||
*, **, *** Significant at p<0.05, <0.01, <0.001; NS, non-significant at p>0.05
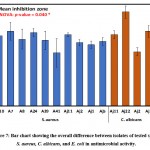 |
Figure 7: Bar chart showing the overall difference between isolates of tested species S. aureus, C. albicans, and E. coli in antimicrobial activity. |
Phylogenetic tree interpretation
The tree was constructed using the unweighted pair-group method with arithmetic mean (UPGMA) method. The evolutionary distances were computed using the Maximum Composite Likelihood method. This analysis involved 10 nucleotide sequences; as there were three identical nucleotides, two were removed, as their pattern of genomic identification with neighboring members was similar. Primarily, the tree comprised two major (18, 16) and six minor (11, 12, 13, 14, 15, 17) clades. The closely related isolates were clustered among the same replicates. Each clade (replicate) was represented by a node ID, and this shows the correlation among the isolates in vicinity. The major clades 18 and 16 comprised ancestral organisms, where clade 16 was branched to form clade 12 and the side branch 6, comprising B. subtilis EBV2 MW512604. Clade 16 was the common group of isolates with ancestral divergence from others. Clade 18 produced two sub-clades (17,15) with each clade representing evolutionary distinct groups. The replicates within clade 18 formed respective sub and minor clades leading to the group of organisms most closely related to node IDs 14, 11, and 15. Similarly, the isolates from node 12 were more closely related and might have common genome attributes.
Discussion
Similar to the methodology of this study, many researchers choose soil as a rich source of microorganisms. Unexplored regions represent a promising reservoir of microbes that are capable of producing antimicrobial substances[18] [10]. Screening for functional antibiotics from natural resources, such as soil, is a well-known approach to discover unique bacterial strains that are capable of producing robust bioactive compounds as a result of their adaptive behavior to compete with other microorganisms in their habitat. Isolated bacteria were characterized and tested for their ability to produce valuable pharmaceutical products. The presence of inhibition zones is an indicator that the isolated soil bacteria are strongly active and produce antimicrobial substances against resistant pathogens that were provided by King Abdulaziz University Hospital.
Recently, resistant bacteria have become prevalent in many countries, and their impact is spreading owing to the excessive use of antimicrobials in hospitals and by the agriculture industry. As this poses a serious threat, it has gained the prominent attention of the WHO and specialized health organizations in countries worldwide. Moreover, the vast majority of research efforts have been devoted to find solutions to reduce the impact of this issue. Therefore, strict actions are required to end the occurrence of resistant strains. The Global Action Plan (GAP) was adopted in 2015 by the World Health Assembly. Among its five strategic goals is to conquer AMR by increasing investments in antibiotic discovery. As a result, the GAP was also adopted by the World Animal Health Organization (OIE) and the Food and Agriculture Organization (FAO)19, 20.
The first antibiotic was discovered by Alexander Fleming in the 1930, and this period is referred to as the “golden era.” Long before the discovery of penicillin, infectious diseases were the leading cause of mortality in the early 1900s 21. The commercialization of penicillin vastly improved the treatment of deadly infectious diseases caused by microbes. Unfortunately, despite the notable effects of penicillin in curing infections, the development of antibiotic resistance followed shortly after 22. The evolution of resistance among a wide range of medically significant pathogenic microorganisms, such as S. aureus, C. albicans, E. coli, and P. aeruginosa, as well as many other species cause recalcitrant infections, which can lead to fatal and incurable diseases 21, 23, 24.
During this study, most isolated bacteria did not restrain the growth of some of the resistant microbes, particularly P aeruginosa (P>0.05), an opportunistic pathogen most commonly associated with nosocomial infections and ventilator-associated pneumonia. This significant finding indicates the potent resistance of this microorganism, as previous studies have shown that P. aeruginosa resistance is multi-factorial, which means it can be manifested by innate, acquired, or adaptive mechanisms. Therefore, the treatment of P. aeruginosa is challenging and requires the development of more powerful and novel antibiotics with different modes of action to overcome multi-drug resistance strains25 . Moreover only four of the 12 strains tested strains inhibited the growth of C. albicans, which is a major causative pathogen responsible for septicemia and a high rate of mortality, as well as nosocomial infections 25, 26. Only one of the 12 strains restrained the growth of E. coli. This low susceptibility leads to great concerns about disseminating resistant genes to other strains via different means without finding an effective strategy to avoid the implications of their virulent traits. Surprisingly, S. aureus showed great susceptibility to the tested antibiotics (P<0.001) despite its high resistance to methicillin.
Similar to our findings, 14 indicated that the genus Bacillus had the highest number of isolates capable of producing antimicrobials against the tested pathogens. Bacillus species are capable of producing antimicrobial polypeptides that can be critical to treat infections. These polypeptides include bacitracin, gramycidin S, polymyxin, and tyrotricidin. In addition, Bacillus antimicrobial activity is known to have a wide spectrum, and thus, it can also be used against fungal infections. Previous studies have shown that Bacillus species antibiotics are more effective against gram-positive bacteria. Our findings demonstrated that all eight strains of Bacillus could inhibit the growth of methicillin-resistant S. aureus 27. Some of these strains were collected from the mangrove rhizosphere. The ecosystem of mangrove trees is a rich environment for microbes to thrive, and therefore, there is a high chance of finding microbial species that produce effective antibiotics in such areas 10. Proper advancement of these technologies and methods to isolate microbes from mangrove ecosystems will enhance the possibility of discovering novel bioactive microbes. Another strongly bioactive microbe, S. venezuelae, was also isolated from Makkah province and tested against the four pathogens. Approximately half of the overall production of antibiotics worldwide is through the isolation of Actinobacteria genera, and in particular Streptomyces. It is the main producer of secondary metabolites with anti-mycobacterial activity (e.g., caprazamycin B, sansanmycins, urdamycinone E, urdamycinone G, dehydroxyaquayamycin, streptcytosine A, chrysomycin A, and dinactin) 3.
Cytobacillus firmus holds great promise as its biosynthesized AgNPs show potent antibacterial and antifungal activities, which were evaluated against S. aureus and E. coli bacteria and pearl millet blast disease-causing fungi Magnoporthe grisea 28 Similarly, in our study, a Cytobacillus spp. exhibited antibacterial activity against S. aureus and antifungal activities against C. albicans. Furthermore, A. veronii is an important pathogen causing freshwater fish sepsis and ulcer syndrome. An increasing number of cases has demonstrated its significance as an aquatic zoonotic agent 29, 30. Therefore, few studies have investigated the biosynthetic capacity of this group of microorganisms. The presence of secondary metabolites has been recognized in various Aeromonas species, including A. veronii 29. In a recent study A. veronii (strain A134) inhibited the growth of Microcystis aeruginosa (strain MGK), and novel secondary metabolites were identified, which yielded, among others, three new metabolites: 9-chlorolumichrome (1), veronimide (2), and veronipyrazine (3)31. In this study A. veronii has shown antibiotic activity against C. albicans, therefore, it proves to have potency against multi-drug resistant microorganisms, despite being one itself.
Conclusion
This research initially aimed to provide compounds to combat resistant pathogens, and different locations were selected to sample soil and isolate microbial organisms. Our results identified a variety of soil microbes that are able to suppress the growth of resistant pathogens, suggesting that soil microorganisms have the ability to overcome resistant bacteria. This work could aid in the discovery of new soil-derived antibiotic-producing microbes using purification, characterization, and structural analyses. Further studies to investigate the quality, mechanisms, and commercial value of these antibiotics are needed.
Acknowledgement
This work was mainly sponsored by Science Research and Innovation Unit, King Abdulaziz University, Jeddah, Saudi Arabia. The research number is (scigriu41/13). The authors also are appreciative to the staff of king Fahed research center and the central labotroy at science collage, king abdulAziz university for their outstanding support.
Conflict of Interest
The authors declare no conflict of interest.
Funding Sources
This work was funded by Mawakeb Alajer Association, Jeddah, Saudi Arabia through the Science Research and Innovation Unit at the Faculty of Science, King Abdulaziz University, Jeddah, Saudi Arabia under research number (scigriu 41/13).
References
- F. (2021) Amábile-Cuevas, “Antibiotic resistance from, and to the environment,” AIMS Environmental Science, vol. 8, no. 1, pp. 18–35, 2021, doi: 10.3934/environsci.2021002.
CrossRef - Ahmad, S. Ahmad, and K. P. Rumbaugh, Antibacterial drug discovery to combat MDR: Natural compounds, nanotechnology and novel synthetic sources. 2019. doi: 10.1007/978-981-13-9871-1.
CrossRef - Abdullah Al-Dhabi, G. Ali Esmail, A. K. Mohammed Ghilan, M. Valan Arasu, and V. Duraipandiyan, “Metabolite profiling of Streptomyces sp. Al-Dhabi-100 isolated from the marine environment in Saudi Arabia with anti-bacterial, anti-tubercular and anti-oxidant potentials,” Journal of King Saud University – Science, vol. 32, no. 2, pp. 1628–1633, Mar. 2020, doi: 10.1016/ j.jksus.2019.12.021.
CrossRef - Al-Amoudi et al., “Bioprospecting red sea coastal ecosystems for culturable microorganisms and their antimicrobial potential,” Marine Drugs, vol. 14, no. 9, Sep. 2016, doi: 10.3390/md14090165.
CrossRef - Ashiru-Oredope et al., “Healthcare workers’ knowledge, attitudes and behaviours with respect to antibiotics, antibiotic use and antibiotic resistance across 30 EU/EEA countries in 2019,” Eurosurveillance, vol. 26, no. 12, Mar. 2021, doi: 10.2807/1560-7917.ES.2021.26.12.1900633.
CrossRef - A. Olowe et al., “Phylogenetic grouping and biofilm formation of multidrug resistant Escherichia coli isolates from humans, animals and food products in South-West Nigeria,” Scientific African, vol. 6, p. e00158, Nov. 2019, doi: 10.1016/j.sciaf.2019.e00158.
CrossRef - Davies and D. Davies, “Origins and Evolution of Antibiotic Resistance,” Microbiology and Molecular Biology Reviews : MMBR, vol. 74, no. 3, p. 417, 2010, doi: 10.1128/MMBR.00016-10.
CrossRef - Liu and M. Pop, “ARDB—Antibiotic Resistance Genes Database,” Nucleic Acids Research, vol. 37, no. Database issue, p. D443, 2009, doi: 10.1093/NAR/GKN656.
CrossRef - Mohsin et al., “Emergence of plasmid-mediated tigecycline resistance tet(X4) gene in Escherichia coli isolated from poultry, food and the environment in South Asia,” Science of the Total Environment, vol. 787, p. 147613, Sep. 2021, doi: 10.1016/j.scitotenv.2021.147613.
CrossRef - Cepas and S. M. Soto, “Relationship between virulence and resistance among gram-negative bacteria,” Antibiotics, vol. 9, no. 10. MDPI AG, pp. 1–11, Oct. 01, 2020. doi: 10.3390/antibiotics9100719.
CrossRef - S. Palla, G. S. Guntuku, M. K. K. Muthyala, S. Pingali, and P. K. Sahu, “Isolation and molecular characterization of antifungal metabolite producing actinomycete from mangrove soil,” Beni-Suef University Journal of Basic and Applied Sciences, vol. 7, no. 2, pp. 250–256, Jun. 2018, doi: 10.1016/j.bjbas.2018.02.006.
CrossRef - Ou et al., “Prevalence of multidrug-resistant Staphylococcus aureus isolates with strong biofilm formation ability among animal-based food in Shanghai,” Food Control, vol. 112, p. 107106, Jun. 2020, doi: 10.1016/j.foodcont.2020.107106.
CrossRef - Oechslin, “Resistance development to bacteriophages occurring during bacteriophage therapy,” Viruses, vol. 10, no. 7. 2018. doi: 10.3390/v10070351.
CrossRef - Tangcharoensathien and P. Travis, “Accelerate Implementation of the WHO Global Code of Practice on International Recruitment of Health Personnel: Experiences From the South East Asia Region ,” Kerman University of Medical Sciences, vol. 5, no. 1, pp. 43–46, 2015, doi: 10.15171/ijhpm.2015.161.
CrossRef - Adeel Rafiq et al., “ISOLATION AND IDENTIFICATION OF ANTIBIOTIC PRODUCING MICROORGANISMS FROM SOIL | INTERNATIONAL JOURNAL OF PHARMACEUTICAL SCIENCES AND RESEARCH,” NTERNATIONAL JOURNAL OF PHARMACEUTICAL SCIENCES AND RESEARCH, 2017, Accessed: Apr. 27, 2021. [Online]. Available: https://ijpsr.com/bft-article/isolation-and-identification-of-antibiotic-producing-microorganisms-from-soil/?view=fulltext
- T. Downs et al., “Volcanic history of the northernmost part of the Harrat Rahat volcanic field, Saudi Arabia,” Geosphere, vol. 14, no. 3, pp. 1253–1282, Jun. 2018, doi: 10.1130/GES01625.1.
CrossRef - R. Moufti, A. M. Moghazi, and K. A. Ali, “40Ar/39Ar geochronology of the Neogene-Quaternary Harrat Al-Madinah intercontinental volcanic field, Saudi Arabia: Implications for duration and migration of volcanic activity,” Journal of Asian Earth Sciences, vol. 62, pp. 253–268, 2013, doi: 10.1016/j.jseaes.2012.09.027.
CrossRef - (2004) Sandle, “Gram ’ s Stain : History and Explanation of the Fundamental Technique of Determinative Bacteriology,” IST Science and Technology Journal, vol. 54, no. March 2004, pp. 3–4, 2004.
- Trenozhnikova and A. Azizan, “Discovery of Actinomycetes from Extreme Environments with Potential to Produce Novel Antibiotics,” Central Asian Journal of Global Health, vol. 7, no. 1, Dec. 2018, doi: 10.5195/CAJGH.2018.337.
CrossRef - H. Organization, “Prioritization of pathogens to guide discovery, research and development of new antibiotics for drug-resistant bacterial infections, including tuberculosis,” World Health Organization, Geneva, 2017. [Online]. Available: https://apps.who.int/iris/handle/10665/311820
- Mattar, S. Edwards, E. Baraldi, and J. Hood, “An overview of the global antimicrobial resistance research and development hub and the current landscape,” Current opinion in microbiology, vol. 57, pp. 56–61, Oct. 2020, doi: 10.1016/J.MIB.2020.06.009.
CrossRef - R. da Cunha, L. P. Fonseca, and C. R. C. Calado, “Antibiotic Discovery: Where Have We Come from, Where Do We Go?,” Antibiotics, vol. 8, no. 2, Jun. 2019, doi: 10.3390/ANTIBIOTICS8020045.
CrossRef - A. Alharbi, M. Wainwright, T. A. Alahmadi, H. bin Salleeh, A. A. Faden, and A. Chinnathambi, “What if Fleming had not discovered penicillin?,” Saudi Journal of Biological Sciences, vol. 21, no. 4, p. 289, 2014, doi: 10.1016/J.SJBS.2013.12.007.
CrossRef - Guo, G. Song, M. Sun, J. Wang, and Y. Wang, “Prevalence and Therapies of Antibiotic-Resistance in Staphylococcus aureus,” Frontiers in Cellular and Infection Microbiology, vol. 10, p. 107, Mar. 2020, doi: 10.3389/FCIMB.2020.00107.
CrossRef - Pang, R. Raudonis, B. R. Glick, T. J. Lin, and Z. Cheng, “Antibiotic resistance in Pseudomonas aeruginosa: mechanisms and alternative therapeutic strategies,” Biotechnology advances, vol. 37, no. 1, pp. 177–192, Jan. 2019, doi: 10.1016/J.BIOTECHADV.2018.11.013.
CrossRef - Sakagami et al., “Antifungal susceptibility trend and analysis of resistance mechanism for Candida species isolated from bloodstream at a Japanese university hospital,” Journal of Infection and Chemotherapy, vol. 25, no. 1, pp. 34–40, Jan. 2019, doi: 10.1016/ J.JIAC.2018.10.007.
CrossRef - H. Lara and J. L. Lopez-Ribot, “Inhibition of Mixed Biofilms of Candida albicans and Methicillin-Resistant Staphylococcus aureus by Positively Charged Silver Nanoparticles and Functionalized Silicone Elastomers,” Pathogens, vol. 9, no. 10, pp. 1–14, Oct. 2020, doi: 10.3390/PATHOGENS9100784.
CrossRef - Yilmaz, H. Soran, and Y. Beyatli, “Antimicrobial activities of some Bacillus spp. strains isolated from the soil,” Microbiological research, vol. 161, no. 2, pp. 127–131, Feb. 2006, doi: 10.1016/J.MICRES.2005.07.001.
CrossRef - Sudarsan et al., “Green Synthesis of Silver Nanoparticles by Cytobacillus firmus Isolated from the Stem Bark of Terminalia arjuna and Their Antimicrobial Activity,” Biomolecules, vol. 11, no. 2, pp. 1–16, Feb. 2021, doi: 10.3390/BIOM11020259.
CrossRef - D. M. Gu, “Antagonism of Bacterial Extracellular Metabolites to Freshwater-Fouling Invertebrate Zebra Mussels, Dreissena polymopha,” Journal of Microbiology, vol. 39, no. 2, pp. 133–138, 2001.
- Li et al., “Aeromonas veronii Infection in Commercial Freshwater Fish: A Potential Threat to Public Health,” Animals : an Open Access Journal from MDPI, vol. 10, no. 4, Apr. 2020, doi: 10.3390/ANI10040608.
CrossRef - Weiss, D. Kovalerchick, O. Murik, A. Sukenik, A. Kaplan, and S. Carmeli, “Secondary Metabolites of Aeromonas veronii Strain A134 Isolated from a Microcystis aeruginosa Bloom,” Metabolites, vol. 9, no. 6, Jun. 2019, doi: 10.3390/METABO9060110.
CrossRef

This work is licensed under a Creative Commons Attribution 4.0 International License.