Manuscript accepted on : 5-Apr-2021
Published online on: 07-04-2021
Plagiarism Check: Yes
Reviewed by: Dr. Robert Susło 
Second Review by: Dr. Yamini Tiwari
Final Approval by: Dr. Imran Ali![]()

Gulsanga Lemar1* , Saleha Shahar1,2
, Saleha Shahar1,2  and Abdul Rahman Osmani3
and Abdul Rahman Osmani3
1Biosciences, Kabul University, Kabul, Afghanistan, and Universiti Teknologi Malaysia
2Biosciences, UniversitiTeknologi Malaysia, Johor Bahru, Malaysia, 81310.
3Zoology, Kabul University, Kabul, Afghanistan, 1001, Afghanistan.
Corresponding Author E-mail: gulsangalemar@gmail.com
DOI : http://dx.doi.org/10.13005/bbra/2905
ABSTRACT:
The purpose of the sewage treatment process is to decrease the concentration of contaminants, including pathogens, before discharging into the receiving streams. And the standard operating procedure of STP in Malaysia is to discharge the treated wastewater with low nutrient and low organic materials into the streams but the bacterial content of the discharge and its risk to the stream’s natural microbial verity or health is unknown. However, studies reported that pathogens could escape sewage treatment plants (STPs) processes and showed health risk of streams impacted by STP effluent. On the other hand, majority of these studies relied on metagenomic strategy, without assessing changes to culturable bacteria. Isolation of living microbes provides realistic risk assessment compared to metagenome survey alone. Therefore, this study aims to determine the presence of culturable pathogenic bacteria from water impacted by STP effluent to establish justifiable public health risk. For that, the presence of bile resistant bacteria was determined from water taken from surface water receiving effluent from STP-1 (Kolej 9, UTM) in Malaysia. Enumeration and isolation of bacteria were done on MacConkey agar through membrane filtration method, followed by partial identification, using Triple Sugar Iron agar (TSI). The result showed that STP effluent changes the diversity, and abundance, of bile resistant bacteria (specifically Enterobacteriaceae family) of receiving streams. Most of the isolated bile resistant bacteria are opportunistic pathogens for human. Findings from this study provide a snapshot of the bigger picture of microbial changes in a stream impacted by STP effluent painted initially by metagenome studies. And shows that despite of treatment, some contaminants (microbes) remained and released into surface waters, which contribute to the water pollutions.
KEYWORDS: Culturable Pathogenic Bacteria; Macconkey Agar; Public Health Risk; Surface Water; Sewage Treatment Plant
Download this article as:| Copy the following to cite this article: Lemar G, Shahar S, Osmani A. R. Influence of sewage treatment plant effluent on the presence of culturable pathogenic bacteria in the water body. Biosci Biotech Res Asia 2021;18(1). |
| Copy the following to cite this URL: Lemar G, Shahar S, Osmani A. R. Influence of sewage treatment plant effluent on the presence of culturable pathogenic bacteria in the water body. Biosci Biotech Res Asia 2021;18(1). Available from: https://bit.ly/3cYTruj |
Introduction
A large portion of water bodies is impacted by different kinds of wastewater released from various sources: household, business properties, industry, and farming 1. Modern wastewater management systems treat the wastewater into low-nutrient and low-organic content for release into the surface water without risk to human wellbeing or harm to the environment.The proficiency of any Sewage Treatment Plant (STP) is specified by the general execution of plant and effluent quality fit from an environmental standpoint 2. In this manner, the system is scrutinized to specify the general pollution associated with it. The efficiency of the system is vulnerable to several factors. Sewage from different sources, such as residential and industrial, produced intricate blends of inorganic and organic constituents causing incompatibility with the system’s operation. The system can also be overwhelmed bythe influent of raw sewage beyond the system’s capacity. High operational cost makes maintenance difficultas well and causes systems to poor performance 3. These threats can reduce the efficiency of the system, causing the release of faecal bacteria, parasites, and viruses with potential health risks such as gastrointestinal and respiratory illness 4.Furthermore, Despite treatment, some contaminants remain in treated wastewater released into surface waters, such as microbes (mostly intestinal) as well as chemicals from personal care items 5. Al-gheethi and Ismail (2014) studied the bacterial assorted variety in treated sewage plus biosolid produced from five STPs in Yemen. 160 bacterial strains were isolated of which, E. coli was the most widely recognized. Osuolale and Okoh (2017) study from the five wastewater treatment plants (WWTPs) in South Africa, Eastern Cape showed that in the treated effluents, the existence of faecal coliforms and E. coli was higher than that of rotaviruses or enteroviruses. Shigellaspp, Salmonellaspp, Staphylococcusspp, Vibriospp, as well as Listeria spp. were isolated from STP in Aswan, Egypt8. Similarly, Pant and Mittal (2007) reported that all three faecal-oral pathogens, Shigella, Salmonella, and Vibrio were notable in all the effluent samples from the plant alongside indicator microorganisms. Their findings recommended that treated sewage routinely contained pathogens as well as faecal coliform (FC) and faecal streptococci (FS).
This demands the question: how do regulated effluent from wastewater treatment plants (STPs) impacts surface water? Metagenomics studies identified changes in the microbial abundance and diversity of surface water that received effluents from STP 10, 11. For instance, the significant increase in abundance of human gut bacteria and decrease of phototrophic microorganisms or even disappearing after mixing upstream and outflow in surface water receiving effluents from (STP) effluent 12. Metagenome is a powerful tool, capable of identifying bacteria, viruses, fungi, and parasites in complex samples through sequencing of DNA fragments. However, the presence of DNA fragments does notguarantee the presence of viable microbes. In this regard, metagenome analyses could not perceive the actual risk of STP effluent on health or the environment.
Unregulated discharges of untreated wastewater are also serious threat causing faecal contamination of surface water. The coliforms are bacteria from the Enterobacteriaceae family resistant to bile to adapt to gut condition. They are common in faecal materials, and can be found in the aquatic environment contaminated with faeces. Because they are easy to grow, and are reliable faecal contamination indicator 13, 14, 15. However, the coliforms are universal faecal bacteria found in all mammals, not only humans. In this sense, they are not effective for differentiating the source of faecal contamination, whether from human such as the STP, runoff from agriculture farms, or even wildlife normally inhabiting near water bodies 16. Recognising the main bacterial residence of the gut or bile-resistant bacteria,the frequently isolated in faecal contaminant can be indicative of potential source indicator 15.
A study showed that even the appropriately treated sewerages are capable of negative ramifications on the self-purification capacities of water reservoirs1. The serious ramifications on human health due to deficient wastewater treatment is underlined by the United Nation: globally, 2 million tons of sewage plus industrial as well as horticultural wastes are released into the world’s waterways. At any time, 1.8 million children under five years of age passed away each year from water-related sicknesses. People who died as a consequence of polluted water exceeded those who perished by all kinds of violence, including wars. A report from the American Academy of Microbiology shows the built-up of global complacency on wastewater treatment could be hazardous; causing widespread sickness every year 16. Therefore, there is an intense need for checking the water quality, the assessment for the presence of infectious bacteria in the water that is harmful to human and animal wellness 17.
This study aims to preliminary assess the effect of STP effluent on surface water by studying the population of culturable gut bile-resistant bacteria in the surface water. Bile-resistant bacteria wereselectively grown on MacConkey agar and partially identified by triple suga iron agar. If bile-resistant bacteria can be isolated from surface water downstream of STP, it is possible the bacteria were faecal-origin and from STP. However, the surface water is open to contamination by animal faeces. Thus, the population of bile-resistant bacteria that closely resembles the population from STP effluent, but different from undisturbed water upstream of STP means that bacteria would likely originate from STP effluent rather than the environment.
Material and methods
Sampling
Samples were collected from the stream receiving effluent of Kolej 9 sewage treatment plant in UniversitiTeknologi Malaysia (UTM) Skudai, Johor Bahru (Figure 1). The stream was sampled at three different points: (1) 5 m upstream from effluent outflow, which does not receive the effluent of STP to determine the presence of microbes in those water, which are not affected by STP effluent (2) effluent outflow, and (3) 5 m downstream of effluent outflow, which can receive the effluent of STP outflow as well as upstream to identify the impact of STP on surface water. Samples were collected by the grab method using a plastic scoop. The scoop was rinsed with water from the sampling site before sample-grab. Water samples were placed in a sterile screw-cap container. Then, the culturing of bacteria was carried out as soon as after sampling.
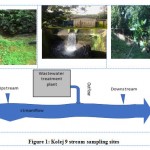 |
Figure 1: Kolej 9 stream sampling sites. |
Detection of Bile Resistance Bacteria in Water Samples
Enumeration of bile resistant bacteria
Isolation plus enumeration of bile resistant bacteria was carried out using standard membrane filtration methods on MacConkey agar (MAC) (Figure 2). MAC agar was used because the incorporation of bile salts and crystal violet make the medium selective towards Gram-negative coliforms and Coccus (bacteria from the guts). The phenol red allows differentiation of lactose fermenting coliform from non-lactose fermenting coliforms. The 50 gr of MacConkey agar powder (Catalogue No: 1.05465.0500 & Brand: Mark KGaA) was dissolved in 1 litre of distilled water using a magnetic stirrer and was autoclaved for 15 minutes at 121°C. The medium was poured into the Petri dishes and solidified at room temperature 18. MacConkey agar plates were labelled with the sample number/identification as well as the sample volume to be analysed. Then, a sterile filter membrane (0.45 μm, Whatman) was placed on the porous plate filter housing (Nalgene) using flamed forceps. The funnel was then attached to the filter unit base 19. Next, 100 ml of the water sample was measured and poured into the funnel. The vacuum pump was switched on for the water sample to pass through the filter membrane. The filter membrane was picked by its side with flamed forceps, gently lifted, and placed face-up on a labelled MacConkey agar plate. To prevent trapping air bubbles between the underlying agar and the filter membrane, the filter was slide onto the agar using a rolling technique. Finally, the agar plate was inverted and incubated at 35°C for 22 to 24 hours. The next day, colonies grown on the filter were counted to find out the bacterial population in surface water. The final values of the colony-forming unit in the water sample were calculated using the following formula: 20
For the diluted samples, the dilution factor was also included as the following equation.
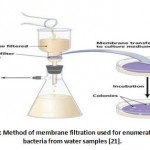 |
Figure 2: Method of membrane filtration used for enumeration of bacteria from water samples [21]. |
Purification of bile resistant bacteria
After the enumeration of bacteria colony-forming units (CFU/100 ml) membrane filter plate, the colonies were differentiated visually according to shape and colour for initial identification. For this, plates with the countable number of colonies used for enumeration, were selected for streaking. Every colony of bacteria on the filter membrane from this countable plate was streaked onto fresh MacConkey agar plates for purification. Each streak plate was labelled and incubated for 22 to 24 hours at 35 °C.
Partial identification of bile resistant bacteria
Bile resistant bacteria growing on MacConkey agar were partially identified using triple sugar iron (TSI) agar (due to the limitation of time). The agar is commonly used to distinguish groups of Enterobacteriaceae, especially for intestinal pathogens based on the ability to ferment carbohydrates and reducing sulphur 22. TSI agar contains glucose, sucrose, and lactose in a concentration of 0.1%, 1%, and 1%, respectively. Phenol red (pH indicator) was used to detect carbohydrate fermentation, which is yellow when below the pH of 6.8. Therefore, the uninoculated medium (pH 7.6) is red from phenol red. In addition, the medium contains two indictors for detecting H2S formation, which are sodium thiosulfate and ferrous sulphate. Thus, it is a two steps process. The H2S is formed from sodium thiosulfate, in the first step. As H2S is a colourless gas, ferrous sulphate, a second indicator is required for visually detecting its production23 . The 65gr of TSI agar (Catalogue No: 1.03915.0500 & Brand: Merck KGaA) was dissolved in 1 litre of distilled water using a magnetic stirrer and was autoclaved for 15 minutes at 121°C. Then, the TSI agar was poured into the sterile universal tubes and was solidified to give agar slant 24.
Next, a small number of bacteria from the 24-hour streak plate was inoculated using the stab and streak inoculation method into the tubes with inoculating wire lope. Then, the tubes were incubated for 22 to 24 hours at 35 °C to identify the opportunistic pathogenic bile resistant bacteria in surface water.
Results
Bile resistant bacteria in surface water before and after receiving STP effluent
The culture was collected twice at the one-month interval (2nd of February and 2nd of March 2020). For the first time the samples, which were collected after rainfall, three different colony colours on MAC were detected after 22 hours to 24 hours incubation at 35°C. The colonies observed were: pink to red, yellow to white, and black(Figure 3).Also, there was less obvious presence of lactose fermenters (pink colonies) in the upstream, and more obvious presence in the downstream. The obvious presence of lactose fermenter colonies in downstream compared to upstream can be an impact of STP outflow, rich in lactose fermenters. Besides, the number of bacterial colonies in outflow was higher than upstream, which caused the downstream to have a high number of bacteria as well (Table 2).
Sampling for the second time was done one-month later, (2nd March 2020). The second time sampling did not have black colonies in any of the samples(Figure 4). As the first-time sapling was done after raining and rainfall can accumulate microbes from the environment, and second time sampling was done when there was no rainfall, thus, rainfall can be the reason for the verity and presence of black colour colonies in the first-time water samples or possibly, the differences between first- and second-time sampling could be caused by changes that happened within one month (interval of two times sampling). Regarding this, studies that monitor the impact of STP on stream showed microbial differences over different times. These studies showed microbes in the stream changed by wetter antecedent moisture conditions, environmental perturbation, physiochemical properties and toxicity of sewage, or hydraulic mixture 25, 26. The change of pattern was high microbial diversity occurring after rainfall, lower microbial diversity after precipitation, and increasing or even disappearing of microorganisms after mixing between upstream and outflow.
In general, culturable bile resistant bacteria in surface water that received STP effluent tend to have majority lactose fermenter. It resembles culturable bile resistant bacteria of STP outflow more than undisturbed upstream.
Partial identification using triple sugar iron agar test
All isolates were partially identified using triple sugar iron (TSI) agar slants incubated for 22 hours to 24 hours at 35°C. Growth on TSI yields nine combinations of characters, (Table 1), and each combination of characters can be attributed to several types of Enterobacteriaceae. The number of times colonies with TSI combination of characters, which were found in enumerated plates, were recorded (Table 2) This qualitative assessment provides a general idea of the occurrence of the types of Enterobacteriaceae in the samples.
Table 2: Possible ID and number of colonies of both time sampling using MacConkey and TSI agar, from upstream, outflow and downstream sampling.
| Presumptive Bacteria ID | Upstream | Outflow | Downstream |
| Number of colonies (10-3) | Number of colonies (10-4) | Number of colonies (10-2) | |
| Citrobacter freundii | 2&2 | 0&1 | 1 & 1 |
| Citrobacter diversus | 0&1 | 0&2 | 0&0 |
| E. coli, E. aerogenes, E. cloacae |
3&36 | 7&64 | 64 &63 |
| Aeromonas hydrophilia | 1&3 | 38& 4 | 22 &0 |
| Alcaligenes faecalis | 6&1 | 5&1 | 1 &0 |
| Serratia, vibrio cholera | 4&13 | 15&10 | 11 &0 |
| Pseudomonas aeruginosa,
pseudomonas putida |
1&1 | 1&0 | 0&0 |
| Klebsiella pneumonia | 4&5 | 2&3 | 4 &1 |
| Shigella dysenteriae, Shigella boydii, Shigella flexneri | 1&4 | 0&0 | 0&0 |
| Salmonella cholerasus, Morganilla morganii | 0&2 | 12&2 | 0&0 |
| Unknown | 3& 3 | 1& 16 | 11& 0 |
| No growth on TSI | 2& 0 | 2 & 5 | 2 & 0 |
| CFU/100 ml (Total) | 3.0 x 103 [27]&7.9×103 [71] | 92×104 [83]&120×104 [108] | 1.29×102 [116]&72×104 [65] |
Growth on TSI showed including some unknown bacteria, Enterobacteriaceae can be found in every sample not only STP outflow but in samples before and after receiving outflow. However, the Enterobacteriaceae population from the sample after STP effluent was introduced into the stream (downstream) was very different from upstream.Outflow from STP changed downstream population to favour 3 to 6 Enterobacteriaceae groups, even though both upstream and outflow almost always carried all 10 members of Enterobacteriaceae. The favoured groups in downstream are Escherichia and Enterobacter, Citrobacter, Klebsiella, Aeromonas, Alcaligenes, and Serratia and Vibrio. Of those favoured, Escherichia and Enterobacter, Citrobacter, and Klebsiella seemed to be a constant feature and Escherichia and Enterobacter as the dominant changes. The abundance of bacterial content (CFU/100 ml) downstream was affected by the entry of outflow into the stream. This is because bacterial count in upstream was3.0x103 CFU/100 ml, very far from the outflow count,which was92x104 CFU/100 ml. On the other hand, bacterial counts downstream were consistently higher,1.29×102 CFU/100 ml.
Discussion
Sewage treatment processes are capable of decreasing the concentration of faecal pathogens 14, 27, 28. However, studies also showed the public health risk of streams impacted by STP effluent as metagenome analyses showed pathogenic bacteria can escape STP treatment processes 29–32. These metagenomic studies found nucleic acid indicators of pathogens such as Bacteroides HF183, Helicobacterspp, E. coli, Enterococci, and Acinetobacter baumannii. In addition, many metagenome studies also showed STP effluent changes the microbial landscape of streams 26, 33,35. And these studies explained changes from the perspective of microbial metabolism. It showed that under long term nutrient stress conditions, such as in wastewater treatment plants, microbial communities developed special metabolic patterns such as specific amino acid metabolism and membrane transporters to maintain optimal cellular activity. However, all of the studies had relied on metagenomic strategy. The very limited study assessed the impact of STP effluent on changes to culturable or living bacteria in the STP and the stream, including pathogenic strains. Isolation of living pathogenic bacteria provide realistic health risk assessment compared to the metagenome survey alone36. Therefore, there is a need to determine the actual presence of culturable bacteria in water impacted by STP effluent to assess any impending public health risk from pathogenic or potentially pathogenic strains. Findings from this study complete the big picture of microbial changes in a stream impacted by STP effluent revealed by metagenome studies and opened up an avenue to potential source-specific bacterial indicators.
In this study, STP effluent (outflow from Kolej 9 STP) was shown to cause the water of the receiving stream to have higher selected groups of Enterobacteriaceae. In addition, the number of bile resistance bacteria in the outflow of STP was higher than upstream, which indicates the presence of bacteria (opportunistic pathogenic bacteria) in the treated wastewater of kolej9 STP and can contribute to the water pollution. The effluent makes the Enterobacteriaceae population in the sample downstream of STP different from that upstream from STP. Particularly, Escherichia (E. coli), Enterobacter (E. aerogenes and E. cloacae), Citrobacter (C. freundii), and Klebsiella (K. pneumoniae) are favoured features. All these are known as opportunistic pathogens for humans. Unlike obligate pathogens, opportunistic pathogens cause infection to those who are immunocompromised either from diseases or poor diet 30. For instance, E. coli is a gut organism. It causes infection of the intestine and causes diarrhoea when contaminated food or water is consumed 37. C. freundii is another intestinal inhabitant of humans, which can be found in environments such as water, sewage, soil, and food. C. freundii may sometimes acquire the ability to produce an enterotoxin, mostly causing abnormal inflammatory changes in the intestinal tract affecting biliary, urinary, and respiratory tracts, and blood of patients with the weak immune system 38. K pneumoniae is present as commensal in the nasopharynx and the intestinal tract. Occasionally, Klebsiella spp. causes human diseases, including asymptomatic colonization of the intestinal, urinary, or respiratory tract, and even fatal septicaemia 38,39. Apart from these favoured feature groups, the presence of Aeromonas hydrophilia and Serratia marcescens or Vibrio cholera, which were also detected in samples, are concerning as these bacteria. Similarly, these bacteria are opportunistic human pathogens. A. hydrophilia causes gastroenteritis, septicaemia, meningitis, and wound infections 39 whereas Serratia marcescens causes respiratory tract infection, urinary tract infection, pneumonia and meningitis 40. Vibrio cholera is responsible for intestinal infections of humans causing cholera worldwide when the bacteria-contaminated drinking water is consumed38.
Does isolation of the mentioned bile resistant bacteria (opportunistic pathogens) imply the health risk of surface water impacted by STP effluent? One of the main bacterial indicators of faecal contamination is Faecal Coliform E. coli. Studies have shown that gastrointestinal and respiratory diseases are linked to polluted waters with high numbers of indicator bacteria16, 41. WHO suggested that Faecal Coliform must be less than 1000 cells/100 ml for harmless recycling of sewage treated effluents 42. The results of the current study showed the presence of opportunistic pathogenic bacteria in the samples taken from the stream impacted by STP and also showed that the total number of E. coli and Enterobacter in downstream is 63×104 CFU/100 ml.
The presence of gut organisms and opportunistic pathogens, such as E. coli, is proof of faecal contamination. However, studies showed E. coli as an indicator of faecal contamination could not tell the source of faecal either from humans or animals. This is because, these indicators are universal faecal indicators found in all mammals, not only humans[16]. Besides, sources of faecal pollution in water varies. For instance, it can be from the human sewage treatment plant, runoff from agriculture farms, or even wildlife that are normal inhabitants around water bodies. [16], [43]. Thus, the inability to differentiate the source of faecal would prevent effective control of faecal pollution. In this study, gut bacteria other than E. coli were also detected in downstream samples, which particularly received the effluent from Kolej9 (a residential place) STP. Partial identification by TSI suggested Enterobacter were very high. Thus, Enterobacter can be a candidate for a source-specific faecal indicator. Common featured bacteria, which are Citrobacter and Klebsiella, or the occasionally detected in high number Serratia, Vibrio, and Aeromonas could be considered as candidates as well.
To date, other studies that researched alternative of E. coli as faecal bacterial indicator had identified bacteria such as Clostridia, Bacteroides, Bifidobacter, enterococci as the possible source-specific faecal indicator 44, 45. However, these bacteria have problems, or their use is limited. There is considerable debate regarding the use of Clostridium perfringens as an indicator of water quality due to its persistence in the environment 46. On the other hand, the need to maintain anoxic conditions for cultivation, isolation, and biochemical identification limits the use of anaerobic Bacteroide species as a faecal indicator 45. Bifidobacterium tolerates some oxygen but is a fastidious bacterium that grows very slowly in culture media, and are the least studied of all faecal bacteria due to the technical difficulties in their isolation and cultivation 39. Several studies have identified difficulties to find media that can efficiently enumerate a wide variety of Enterococcus spp. Not sacrificing the specificity of the Enterococcus genus and the detection of enterococci isolates from environmental matrices (e.g. sediments, soil, sand, plants, plus water) remains challenging 47]. However, it is still too early to suggest bacteria as alternatives to E. coli but, as such bacteria cannot be definitive proof of faecal contamination. Results from current work open up the possibility of other possible source-specific faecal indicator candidates that can be further researched.
Furthermore, the characteristics of the ideal indicator organism are: 1. suitable for all types of water. 2. Present in greater quantities than pathogens. 3. Present in sewage 4. They should be at least as resistant as the pathogen to environmental threats and disinfection processes of wastewater treatment plants. 5. The indicator organism should be non-pathogenic. 6. Occur in large numbers in the intestine and faeces. 8. Simple, accurate, and cheap to observe and enumerate. Not multiplying outside the enteric environment is the desired character as well 39, 45, 47, 48. A perfect organism with all the criteria does not exist. Even existing faecal coliform E. coli is having concern with replication in the environment 49. But studies that focused on the viable count of faecal or gallbladder bacteria from pig, human, and poultry sources, found the E. coli as the majority and most abundant across the different sources. Accompanying the E. coli, other bacteria such as Pseudomonas and Aeromonas were easily found in poultry 50, Enterobacter for humans[51], and Salmonella for pigs52. In this study, E. coli and Enterobacter were also found in favour of samples related to STP effluent, instead of the upstream sample without effluent. Perhaps a consortium of faecal bacteria, instead of a single type of coliform, is the way to go for source tracking.
Furthermore, the black colour colony of bacteria on MacConkey agar, found in first time sampling of the current study are the colony, which is not reported about in the previous studies thus, it can provide an avenue for other researchers to do further researches to find about the risk or usefulness of this black colour colony of bacteria on MacConkey agar.
Conclusion
The characteristics of treated sewage for discharge according to Malaysia Standard are low nutrient substances and organic materials. The coliform count is not included. Thus, it is not clear how Malaysia Standard comply STP effluent would affect bacterial diversity and health safety of surface water. This study showed that bile resistance bacteria were high in surface water that received STP effluent than upstream, which does not receive effluent from STP. In addition, STP effluent increased faecal-related Enterobacteriaceae in the surface water. The Enterobacteriaceae are also known to be opportunistic pathogenic bacteria. The presence of culturable opportunistic pathogenic bacteria could be a concern of public health risk. Besides, the detection of opportunistic pathogens in the wastewater of this research would facilitate decision-making for effective technology and management solutions to decrease microbial risks in receiving water bodies. Thus, further research and additional treatment are required to improve the treatment process and reduce the concentration of pathogens in treated sewage effluents. Additionally, this study found that STP effluents contain bile resistance bacteria associated with the human that can be suitable as a source-specific faecal indicator for human sewage.
Acknowledgement
This study was made possible through a post-graduate scholarship funded by the Ministry of Higher Education Afghanistan and Higher Education Development Program (HEDP), and Faculty of Science, Universiti Teknologi Malaysia (UTM) for research facilities and amenities.
Conflict of Interest
No conflict of interest
Funding Source
No sources
References
- Babko, T. Kuzmina, Z. Suchorab, M. K. Widomski, and M. Franus, “Influence of Treated Sewage Discharge on the Benthos Ciliate Assembly in the Lowland River,” Ecol. Chem. Eng. S, vol. 23, no. 3, pp. 461–471, Sep. 2016, doi: 10.1515/eces-2016-0033.
CrossRef - R. Gedekar, M. T. Scholar, E.- Scientist, M. P. Bhorkar, P. K. Baitule, and M. T. Scholar, “Performance Evaluation of Sewage Treatment Plant ( STP ) – A Review,” Int. J. Sci. Technol. Eng. |, vol. 2, no. 07, pp. 2011–2013, 2016.
- N. Edokpay, J. O. Odiyo, and O. S. Durowoju, “Impact of Wastewater on Surface Water Quality in Developing Countries: A Case Study of South Africa,” in School of Enviromental Sciences, 2012, pp. 401–416.
- Y. Lim, S. L. Ong, and J. Hu, “Recent advances in the use of chemical markers for tracing wastewater contamination in aquatic environment: A review,” Water (Switzerland), vol. 9, no. 2, p. 26, 2017, doi: 10.3390/w9020143.
CrossRef - N. Wakode and S. U. Sayyad, “Performance Evaluation of 25MLD Sewage Treatment Plant ( STP ) at Kalyan,” Am. J. Eng. Res., vol. 03, no. 03, pp. 310–316, 2014.
CrossRef - A. S. Al-gheethi and N. Ismail, “Biodegradation of Pharmaceutical Wastes in Treated Sewage Effluents by Bacillus subtilis 1556WTNC,” Env. Process, vol. 1, pp. 459–481, 2014, doi: 10.1007/s40710-014-0034-6.
- Osuolale and A. Okoh, “Human enteric bacteria and viruses in five wastewater treatment plants in the Eastern Cape, South Africa,” J. Infect. Public Health, no. 676, p. 7, 2017, doi: 10.1016/j.jiph.2016.11.012.
CrossRef - Younis, H. A. Soleiman, and K. A. Elmagd, “Microbiological and Chemical Evaluation of Bentonite as a New Technique for Sewage Water Treatment, Aswan City , Egypt,” Seventh Int. Water Technol. Conf. Egypt 1-3, pp. 323–334, 2003.
- Pant and A. K. Mittal, “Monitoring of Pathogenicity of Effluents from the UASB Based Sewage Treatment Plant,” Env. Monit Assess, vol. 133, pp. 43–51, 2007, doi: 10.1007/s10661-006-9558-1.
CrossRef - Chaudhary, I. Kauser, and A. Ray, “Taxon-Driven Functional Shifts Associated with Storm Flow in,” Appl. Environ. Sci., vol. 3, no. 4, pp. e00194-18, 2018.
CrossRef - Garcia, N. Brion, and P. Servais, “Seasonal Variations and Resilience of Bacterial Communities in a Sewage Polluted Urban River,” PLoS One, vol. 9, no. 3, p. e92579, 2014, doi: 10.1371/journal.pone.0092579.
CrossRef - Clinton et al., “Sediment Microbial Diversity in Urban Piedmont North Carolina Watersheds Receiving Wastewater Input,” water, vol. 12, no. 6, p. 1557, 2020.
CrossRef - Dhakal and N. Roshan, “Microbiological quality of slaughterhouses and antibiotic susceptibility pattern of some isolates,” Science (80-. )., pp. 1–52, 2014, doi: 10.13140/RG.2.1.3819.3689.
- Hendricks and E. J. Pool, “The effectiveness of sewage treatment processes to remove faecal pathogens and antibiotic residues,” J. Environ. Sci. Heal. – Part A Toxic/Hazardous Subst. Environ. Eng., vol. 47, no. 2, pp. 289–297, 2012, doi: 10.1080/10934529.2012.637432.
CrossRef - Ivaylo, Y. Todorova, L. Kenderov, and Y. Topalova, “Assessment of Contamination With Opportunistic Pathogenic Bacteria From Family Enterobacteriaceae in Sediments of Iskar River,” Ecol. Eng. Environ. Prot., no. IX, pp. 47–55, 2017.
- M. Monsalvo, “Wastewater and Public Health: Bacterial and Pharmaceutical Exposures,” in Wastewater and Public Health, 2015, pp. 3–279.
CrossRef - Páll, M. Niculae, C. D. Şandru, and M. Spînu, “Human impact on the microbiological water quality of the rivers,” J. Med. Microbiol., vol. 62, no. PART 11, pp. 1635–1640, 2013, doi: 10.1099/jmm.0.055749-0.
CrossRef - Ji and Y. tin Wang, “Selenium reduction by a defined co-culture of Shigella fergusonii strain TB42616 and Pantoea vagans strain EWB32213-2,” Bioprocess Biosyst. Eng., vol. 42, no. 8, pp. 1343–1351, 2019, doi: 10.1007/s00449-019-02134-5.
CrossRef - Oshiro, “Method 1604: Total Coliforms and Escherichia coli in Water by Membrane Filtration Using a Simultaneous Detection Technique (MI Medium).,” United States Environ. Prot. Agency, p. Washington, DC 20460, 2002, [Online]. Available: http://www.epa.gov/nerlcwww/1604sp02.pdf.
- S. and T. U.S. EPA Office of Water, “Method 1604 : Total Coliforms and Escherichia coli in Water by Membrane Filtration Using a Simultaneous Detection Technique ( MI Medium ),” 2002, pp. 1–14.
- Kenneth, “Seasonal prevalence of faecal indicators and enteric pathogens in Suva lagoon,” 2011.
- Y. Aditi, S. S. Rahman, and M. Hossain, “A Study on the Microbiological Status of Mineral Drinking Water,” Open Microbiol. Journal, vol. 11, pp. 31–44, 2017, doi: 10.2174/1874285801711010031.
CrossRef - K. O. M. Paníková, Special Bacteriology Basic Laboratory Test. 2016.
- T. Aung and P. P. Oo, “Isolation and Characterization of Rhizobium From Root Nodules of Arachis Hypogaea L . ( Groundnut ),” J. Myanmar Acad. Arts Sci., vol. XVIII, no. 4, pp. 197–210, 2020.
- Tryland et al., “Impact of rainfall on microbial contamination of surface water,” Int. J. Clim. Chang. Strateg. Manag., vol. 3, no. 4, pp. 361–373, 2011.
CrossRef - Li, X. Jiang, J. Wang, K. Wang, and B. Zheng, “Effect of Sewage and Industrial Effluents on Bacterial and Archaeal Communities of Creek Sediments in the Taihu Basin,” water, vol. 9, no. 373, pp. 1–19, 2017, doi: 10.3390/w9060373.
CrossRef - Drury, E. Rosi-Marshall, and J. J. Kelly, “Wastewater Treatment Effluent Reduces the Abundance and Diversity of Benthic Bacterial Communities in Urban and Suburban Rivers,” Appl. Environ. Microbiol., vol. 79, no. 6, pp. 1897–1905, 2013, doi: 10.1128/aem.03527-12.
CrossRef - Henrique and O. Dias, “Bacteriophages as surrogates of viral pathogens in wastewater treatment processes,” 2016.
- Ahmed, S. Payyappat, M. Cassidy, and C. Besley, “Enhanced insights from human and animal host-associated molecular marker genes in a freshwater lake receiving wet weather overflows,” Sci. Rep., no. 9, pp. 1–13, 2019, doi: 10.1038/s41598-019-48682-4.
CrossRef - Cui and S. Liang, “Monitoring Opportunistic Pathogens in Domestic Wastewater from a Pilot-Scale Anaerobic Biofilm Reactor to Reuse in Agricultural Irrigation,” water, vol. 11, no. 1283, pp. 1–14, 2019.
CrossRef - Hembach, J. Alexander, C. Hiller, A. Wieland, and T. Schwartz, “Dissemination prevention of antibiotic resistant and facultative pathogenic bacteria by ultrafiltration and ozone treatment at an urban wastewater treatment plant,” Sci. Rep., no. 9, pp. 1–12, 2019, doi: 10.1038/s41598-019-49263-1.
CrossRef - Numberger et al., “Characterization of bacterial communities in wastewater with enhanced taxonomic resolution by full-length 16S rRNA sequencing,” Sci. Rep., vol. 9, pp. 1–14, 2019, doi: 10.1038/s41598-019-46015-z.
CrossRef - R. Price, S. H. Ledford, M. O. Ryan, L. Toran, and C. M. Sales, “Wastewater treatment plant effluent introduces recoverable shifts in microbial community composition in receiving streams,” Sci. Total Environ., vol. 613–614, pp. 1104–1116, 2018, doi: 10.1016/j.scitotenv.2017.09.162.
CrossRef - Yang, L. Wang, F. Xiang, L. Zhao, and Z. Qiao, “Activated Sludge Microbial Community and Treatment Performance of Wastewater Treatment Plants in Industrial and Municipal Zones,” Environ. Res. Public Heal., vol. 17, no. 436, pp. 1–15, 2020.
CrossRef - Michael et al., “Trace levels of sewage effluent are sufficient to increase class 1 integron prevalence in freshwater biofilms without changing the core community,” Water Res., vol. 106, pp. 163–170, 2016, doi: 10.1016/j.watres.2016.09.035.
CrossRef - Ben Maamar et al., “Mobilizable antibiotic resistance genes are present in dust microbial communities,” PLOS Pathog., pp. 1–21, 2020.
CrossRef - Makvana and L. R. Krilov, “Escherichia coli Infections,” Am. Acad. Pediatr., vol. 36, no. 4, pp. 167–171, 2015, doi: 10.1542/pir.36-4-167.
CrossRef - M. Amin, “Perspectives on Gastro-Intestinal Pathogenic Bacteria Infections in Humans,” EC Microbiol., vol. 8, no. 11, pp. 1173–1185, 2019.
- Cabral, “Water microbiology. Bacterial pathogens and water,” Int. J. Environ. Res. Public Health, vol. 7, no. 10, pp. 3657–3703, 2010, doi: 10.3390/ijerph7103657.
CrossRef - T. Rudhy, “Isolation , Identification and Molecular Characterization of Pathogenic Organisms Obtained from Meat samples ( Cooked , Semi- cooked and Raw ) of Different Areas of Dhaka City,” 2017.
- F. Arnold et al., “Original Contribution Acute Illness Among Surfers After Exposure to Seawater in Dry- and Wet-Weather Conditions,” Am J Epidemiol, vol. 186, no. 7, pp. 866–875, 2017, doi: 10.1093/aje/kwx019.
CrossRef - Jeong, H. Kim, and T. Jang, “Irrigation Water Quality Standards for Indirect Wastewater Reuse in Agriculture : A Contribution toward Sustainable Wastewater Reuse in South Korea,” WATER, vol. 8, no. 169, pp. 1–18, 2016, doi: 10.3390/w8040169.
CrossRef - W. S. Domingo and T. A. Edge, “Identification of primary sources of faecal pollution,” in Safe Management of Shellfish and HarvestWaters, 2010, pp. 52–80.
- Saeidi et al., “Occurrence of Traditional and Alternative Fecal Indicators in Tropical Urban Environments under Different Land Use,” Appl. Environ. Microbiol., vol. 84, no. 14, pp. 1–16, 2018.
CrossRef - Saxena, R. N. Bhargava, and A. Raj, “Microbial indicators , pathogens and methods for their monitoring in water environment,” J. Water Health, vol. 13, no. 2, pp. 319–339, 2015, doi: 10.2166/wh.2014.275.
CrossRef - M. Scott, J. B. Rose, T. M. Jenkins, and S. R. Farrah, “Microbial Source Tracking : Current Methodology and Future Directions †,” Appl. Environ. Microbiol., vol. 68, no. 12, pp. 5796–5803, 2002, doi: 10.1128/AEM.68.12.5796.
CrossRef - N. Byappanahalli et al., “Enterococci in the Environment,” Microbiol. Mol. Biol. Rev., vol. 76, no. 4, pp. 685–706, 2012, doi: 10.1128/MMBR.00023-12.
CrossRef - D. N. Myers, D. M. Stoeckel, R. N. Bushon, D. S. Francy, and A. M. G. Brady, “Fecal Indicator Bacteria,” in Biological Indicators, vol. 1, 2015, pp. 5–73.
- J. Horan, “Faecal indicator organisms,” in Handbook of Water and Wastewater Microbiology, Elsevier, 2003, pp. 105–112.
CrossRef - Ali, T. Molla, S. Mahmud, K. A. Talukder, and A. K. M. Mohiuddin, “PTC & B Therapeutic Potential of Plant Extracts Against Multidrug Resistance Poultry Bacteria,” Plant Tissue Cult. Biotechnol., vol. 30, no. 1, pp. 119–130, 2020.
CrossRef - D. Perry et al., “Prevalence of faecal carriage of Enterobacteriaceae with NDM-1 carbapenemase at military hospitals in Pakistan, and evaluation of two chromogenic media,” J. Antimicrob. Chemother., vol. 10, no. 66, pp. 2288–2294, 2011, doi: 10.1093/jac/dkr299.
CrossRef - Evangelopoulou, G. Filioussis, and S. Kritas, “Isolation and Antimicrobial Testing of Aeromonas spp ., Citrobacter spp ., Cronobacter spp ., Enterobacter spp ., Escherichia spp ., Klebsiella spp ., and Trabulsiella spp . from the Gall … Isolation and Antimicrobial Testing of Aeromonas spp ., Citroba,” Polish J. Microbiol., vol. 2, no. 64, pp. 185–188, 2017.
CrossRef
(Visited 463 times, 1 visits today)

This work is licensed under a Creative Commons Attribution 4.0 International License.










