Manuscript accepted on : 16-12-2020
Published online on: 18-01-2021
Plagiarism Check: Yes
Reviewed by: Dr. Nagendra Chandrawanshi
Second Review by: Dr. Kulvinder Kaur
Final Approval by: Dr. Alexsandro Galdino![]()
Molecular Fingerprinting of Indian Medicinal Tree Sara Asoca using RAPD Markers
Shailendra Singh Yadav  and Ashwini A. Waoo
and Ashwini A. Waoo
Department of Biotechnology, Faculty of Life Sciences and Technology, AKS University, SATNA, MP, India
Corresponding Author E-mail: biotech.yadav0@gmail.com
DOI : http://dx.doi.org/10.13005/bbra/2874
ABSTRACT:
Saraca asoca is an important medicinal tree facing a serious problem of reduction from its instinctivetenancy in India.Before formulation of conservation strategies for geographical protection of S. asoca genotypes available in India, it is necessary to characterize them. In the current study, the RAPD markers have been utilized effectively for categorization of S. asoca collected from 15 diverse sites of India. An initial experiment on the amplification suitability of genomic DNA samples of four S. asoca was done with 35RAPD primers. Among them only twenty sixproved their efficiency in two times repeat amplification.Total 146 bands were amplified and out of these 97 bands were found to be polymorphic. The average numbers of total band was 5.61 while average numbers of polymorphic bands was 3.73. The numbers of bands produced per primer ranged from 3 (OPE-15) to 8 (RUF205). Among all studied markers the highest percentage (100%) of polymorphism was demonstrated by only one marker (OPE-06). The lowest percent of polymorphicm (20%) was demonstrated by marker RUF211. The average percentage of polymorphism was 66.44%. Cluster analysis grouped all the S. asoca genotypes under study into two groups. Grouping of genotypes according to their sites of collection demonstrates higher similarity among or between them. The results obtained in the current study may help to formulate conservation strategies for the conservation of S. asoca genotypes.
KEYWORDS: Conservation; Fingerprinting; Genotypes; Molecular Markers; Polymorphism
Download this article as:| Copy the following to cite this article: Yadav S. S, Waoo A. A. Molecular Fingerprinting of Indian Medicinal Tree Sara Asoca using RAPD Markers. Biosci Biotech Res Asia 2020;17(4). |
| Copy the following to cite this URL: Yadav S. S, Waoo A. A. Molecular Fingerprinting of Indian Medicinal Tree Sara Asoca using RAPD Markers. Biosci Biotech Res Asia 2020;17(4). Available from: https://bit.ly/2M0RfaE |
Introduction
Molecular fingerprinting technique has been applied enormously for characterization of various medicinal plants, crops as well as tree species (Tripathi et al. 2018). In contrast to morphological markers, DNA based markers are not depends on environmental changes. For proper credentialsof closely related botanicals, it is important to use molecular markers. Saraca asoca (Roxb.) De Wilde, (Family: Caesalpiniaceae) is an important medicinal tree facing a serious problem of reduction from its instinctivetenancy in India. It is now categorized as ‘vulnerable’ and considered ‘red listed’ by the International Union for Conservation of Nature (IUCN) (Senapati et al. 2012; Mohan et al. 2017). In terms of remedial significance of S. asoca, diverse plant parts (seeds, leaves, bark and flowers) have been found better and are in use for the development of different formulations (Hegde et al. 2017).
Among all molecular markers Random amplified polymorphic DNA (RAPD) has been found to bewell-situated in concert which does not necessitate any former information of DNA targeted for the investigation. The RAPDs have proved their significance for inherent multiplicity estimation in various plant species (Tripathi et al. 2013, Khare et al. 2013). Before formulation of conservation strategies for geographical protection of S. asoca genotypes available in India, it is necessary to characterize them. Molecular characterization of S. asoca genotypes have not been performed in at big level in India. So, the current study demonstrates the effective application of RAPD markers for rapid cataloging of fifteen S. asoca genotypes collected from diverse parts of India.
Materials and methods
Plant material
Young leaves of the collected S. asoca genotypes were taken for genomic DNA extraction. An inclusive detail of collected samples with their collection sites listed (Table 1).
Table 1: Details of collection site of Saracaasoca genotypes.
| S. no. | Place of collection | State | Latitude | Longitude |
| 1 | Kodaikanal-1 | SA1 | Tamil Nadu | 10° 15′ N |
| 2 | Kodaikanal-2 | SA2 | Tamil Nadu | 10° 15′ N |
| 3 | Melpallum | SA3 | Tamil Nadu | 10° 20′ N |
| 4 | Palani | SA4 | Tamil Nadu | 10° 26′ N |
| 5 | Satyamagalum-1 | SA5 | Tamil Nadu | 11° 29′ N |
| 6 | Satyamagalum-2 | SA6 | Tamil Nadu | 11° 29′ N |
| 7 | Satyamagalum-3 | SA7 | Tamil Nadu | 11° 29′ N |
| 8 | Satyamagalum-4 | SA8 | Tamil Nadu | 11° 29′ N |
| 9 | Munnar road forest area-1 | SA9 | Kerala | 10° 11′ N |
| 10 | Munnar road forest area-2 | SA10 | Kerala | 10° 11′ N |
| 11 | Gopal swami hill | SA11 | Karnataka | 11°12 N |
| 12 | Borivali forest area | SA12 | Maharashtra | 19 ° 14′ N |
| 13 | Vasco | SA13 | Goa | 15 ° 24′ N |
| 14 | Veterinary College, Jabalpur | SA14 | Madhya Pradesh | 23 ° 06′ N |
| 15 | Gwarighat, Jabalpur | SA15 | Madhya Pradesh | 23 ° 06′ N |
Methods
Genomic DNA was extracted following the steps involved in CTAB method (Doyle and Doyle, 1990). Extracted DNA was purified with RNase treatment. 1X TE buffer was used to dissolve the dried pellet. Dissolved DNA samples were stored at -20 ºC for further use. DNA was quantitatively estimated by UV-spectrophotometer at 260 nm. Decamer (RAPD) primers (Table 2) were used to standardize PCR conditions. DNA templates were amplified with thermal cycler in a total reaction of 20 µl contained template DNA (50 ng), MgCl2 (50 mM), primer (0.4mM), each dNTP (2 mM) and Taq Polymerase (1U) following initial denaturation (5 min at 94 ºC) at first step, followed by denaturation (1 min at 94 ºC), annealing (1 min at 37 ºC) and extension (2 min at 72 ºC) in second step of 45 cycles. In third step final extension (10 min at 72 ºC) was performed. Amplicons were electrophoresed on 1.5% agarose gel in TAE buffer and images were captured under gel documentation system.
Table 2: Sequences of RAPD Primers used in the Study.
| S. | Primer | 5’-3’ Sequence | GC content | Total Bands | Polymorphic Bands | Percentage
Polymorphism |
| 1. | OPA-5 | AGGGGTCTTG | 60% | 8 | 3 | 37.50 |
| 2. | OPA-8 | GTGACGTAGG | 60% | 6 | 4 | 66.60 |
| 3. | OPC-10 | TGTCTGGGTG | 60% | 6 | 5 | 83.30 |
| 4. | OPC-15 | GACGGATCAG | 60% | 5 | 4 | 80.00 |
| 5. | OPAP-07 | ACCACCCGCT | 70% | 5 | 3 | 60.00 |
| 6. | OPAP-13 | TGAAGCCCCT | 60% | 7 | 5 | 71.42 |
| 7. | OPR-15 | GGACAACGAG | 60% | 6 | 5 | 83.30 |
| 8. | OPM 05 | GGGAACGTGT | 60% | 7 | 4 | 57.10 |
| 9. | OPM-06 | CTGGGCAACT | 60% | 7 | 5 | 71.42 |
| 10. | OPM 13 | GGTGGTCAAG | 60% | 6 | 5 | 83.30 |
| 11. | OPO-20 | ACACACGCTG | 60% | 6 | 5 | 83.30 |
| 12. | OPB-18 | CCACAGCAGT | 60% | 5 | 4 | 80.00 |
| 13. | OPE-06 | AAGACCCCTC | 60% | 4 | 4 | 100.0 |
| 14. | OPE-15 | ACGCACAACC | 60% | 3 | 2 | 66.60 |
| 15. | RUF 202 | TTGGCGGCCT | 70% | 4 | 3 | 75.00 |
| 16. | RUF 205 | TGGGTCCCTC | 70% | 3 | 2 | 66.60 |
| 17. | RUF 207 | CAGGCCCTTC | 70% | 6 | 5 | 83.30 |
| 18. | RUF 210 | TGCCGAGCTG | 70% | 5 | 4 | 80.00 |
| 19. | RUF 211 | GGGTAACGCC | 70% | 5 | 1 | 20.00 |
| 20. | RUF 215 | GCTGCGTGAC | 70% | 6 | 2 | 66.60 |
| 21. | RUF 216 | CAGCGAACTA | 50% | 7 | 2 | 28.57 |
| 22. | RUF 217 | CGACTCACAG | 60% | 5 | 4 | 80.00 |
| 23. | RUF 218 | GGGCCTCTAT | 60% | 5 | 4 | 80.00 |
| 24. | RUF 219 | CTAGAGGTCC | 60% | 6 | 4 | 66.6. |
| 25. | RUF 220 | GGGTGAACCG | 70% | 6 | 3 | 50.00 |
| 26. | OPO-03 | TCCGATGCTG’ | 60% | 7 | 5 | 71.42 |
| Total | 146 | 97 | 66.44 | |||
| Average | 5.61 | 3.73 | — | |||
Data analysis
Amplified bands were scored according to their absence ‘0′ or presence ‘1′. The clustering of genotypes was performed on the basis of Jaccard’s similarity coefficient. The dendrogram was constructed by using the UPGMA cluster analysis with help of software NTSYS-pc ver. 2.0 (Rohlf, 2000).
Results and Discussion
Assessment of inherent dissimilarity is very of the essence for the continuance of plant genetic resources in their native territory (Khare et al. 2013). An Initial experiment on the amplification suitability of genomic DNA samples of four S. asoca was done with 35 RAPD primers. Among them only twenty six primers (Table 2) proved their efficiency of the amplification ofconsistentsharp bands.These efficient markers were further used for the amplification of DNA of all studied genotypes. Banding pattern of OPA-05 primer is illustrated in fig. 1. During the present study, total 146 bands were amplified and out of these 97 bands were found to be polymorphic and 49 bands were monomorphic. The average numbers of total band was 5.61 while average numbers of polymorphic bands was 3.73. The number of bands produced per primer ranged from 3 (OPE-15) to 8 (RUF205). Among all studied markers the highest percentage (100%) of polymorphism was demonstrated by only one marker (OPE-06). However rest of the markers had less polymorphism comparatively. The lowest percent of polymorphism (20%) was demonstrated by marker RUF211. The average percentage of polymorphism was 66.44%.
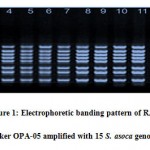 |
Figure 1: Electrophoretic banding pattern of RAPD marker OPA-05 amplified with 15 S. asoca genotypes |
Cluster analysis was performed on the basis of Jaccard’s similarity co-efficient generated from RAPD fingerprinting. The clusteranalysis grouped all the S. asoca genotypes under study into two groups i.e. group A and group B (Fig. 2). Group A is a minor cluster consisting only twoS. asoca genotypes namely SA-9 and SA-10. Both of these genotypes were collected from Munnar road forest area, Kerala. Genotypes SA-9 and SA-10 showed higher similarity and group together.
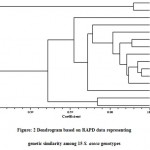 |
Figure: 2 Dendrogram based on RAPD data representing genetic similarity among 15 S. asoca genotypes |
Group B is a major cluster consisting 13 genotypes. Group B was further divided into two sub groups C and D. Sub group C consisted 11 genotypes namely SA-11, SA-4, SA-1, SA-2, SA-3, SA-6, SA-8, SA-7, SA-5, SA-12 and SA-13. Among these eleven genotypes, SA-5 and SA-7 had maximum similarity however SA-13 showed highest genetic divergence from other ten genotypes of sub group C. This genotype was collected from Goa. Eight genotypes collected from different locations of Tamil Nadu state and one genotype from Karnataka, one from Maharashtra and one from Goa are also present in this sub group.The clustering of genotypes in sub group D demonstrated higher similarity among the genotypes collected from Tamil Nadu. However, the genotypes collected from Maharashtra, Goa and Karnataka showed genetic distance from Tamil Nadu genotypes and were clustered distantly. Further, sub group D had only two genotypes coded as SA-14 and SA-15. Both of these genotypes were collected from Jabalpur district of Madhya Pradesh. Geographically, secluded genotypes are tending to build up genetic variability throughout the way of ecological adjustments. The current study is an effort to ascertain the inherent multiplicity backdrop in S. asoca with the use of RAPD markers. Moderate levels of polymorphism observed in the current examination divulge that RAPD technique proved its suitabilityfor fingerprinting studies. The results obtained under this study will lay concrete on the wayfor comprehensiveexploration to recognize all the facets of thisdiscrepancy. RAPD technique has been used extensively for thedetection of diversity among and between medicinal plants (Tripathi et al.2012; Pagare et al. 2017).
Among all DNA fingerprinting techniques RAPDmarker technique is one of the easiest and cheaperthan other techniques available. Because, RAPD markers are decamer and produce multiple bands targeting multi locus in the genome of objective genotype (Khare et al. 2013). The current study demonstrates the suitability of RAPD markers for genetic diversity analysis at DNA level. This particular technique has been employed all the time more formolecular characterization studies in different plant species (Tripathi et al. 2013) and it provides helpful figures on assortment all the way through their aptitude to perceive variations at the DNA level.
Acknowledgement
Author is very thankful to Scientist Dr. S.K. Tiwari Branch Head Forest Genetics, Plant Propagation and Biotechnology who provided all lab facility and time to time he gave me his valuable scientific advised and helped me get results of better quality. I would like to express my special thanks of gratitude to my dear Director, State Forest Research Institute Dr. G. Krishnamurthy who supported encouraging my research work and give new innovative ideas in scientific manner. I also thankful to Dr. Dharmendra Verma who permitted as much as research works had done by me. I can use it in my Ph.D degree. We also thankful to AKS University Department of Biotechnology who give research platform.
Conflict of Interest
Authors have declared no conflicts of interests.
Funding source
The fund given by the National Medicinal Plant Board New Delhi, India to State Forest Research Institute, Jabalpur of this research work
References
- Tripathi N, Shrivastava D, Mir B. A, Kumar S, Govil S, Vahedi M and Bisen P. S. Metabolomic and biotechnological approaches to determine therapeutic potential of Withaniasomnifera(L.) Dunal: A review. , 2018; 50:127-136.
CrossRef - Senapati S. K, Das G. K, Aparajita S and Rout G. R. Assessment of geneticvariability in the Asoka Tree of India. Biodivers., 2012, 13(1), 16–23.
CrossRef - Mohan C, Reddy M. S, Naresh B, Kumar S. M, Fatima S, Manzelat B and Cherku, P. D. RAPD studies of Saraca asoca by fluorescent-labeled primers and development of micropropagation protocol for its conservation. Int. J. Appl. Agri. Res., 2017; 12(2): 137–151.
- Hegde S, Hegde H. V, Jalalpure S. S, Peram M. R, Pai S. R and Roy S. Resolving identification issues of Saraca asoca from its adulterant andcommercial samples using phytochemical markers. Pharmacog. Magaz., (2017); 13(Suppl. 2) S266.
CrossRef - Tripathi N, Saini N and Tiwari S. Morphological and molecular characterization of endangered medicinal plant species Coleus forskohlii collected from central India. J. Crop Sci. Biotechnol., 2013; 16(4): 253- 261.
CrossRef
- Khare D, Bisen A, Nair P and Tripathi N. Genetic diversity in soybean germplasm identified by RAPD markers. Asia Pac. J. Mo. Biol. Biotechnol., 2013; 21: 121-123.
- Doyle J. J and Doyle J. L. Isolation of plant DNA from fresh tissue. Focus, 1990; 12, 3-15.
- Rohlf F. J. NTSYS-pc: numerical taxonomy system ver.2.1.Exeter Publishing Ltd, Setauket. 2002.
- Tripathi N, Saini N, Kumar S and Tiwari S. Assessment of Genetic Diversity among WithaniasomniferaCollected from Central India using RAPD and ISSR Analysis. J. Med. Arom. Plant Sci. Biotechnol., 2012; 6(1): 133-139
- Pagare S, Mishra R. P, Tripathi N, Rathore M and Kumar B. Assessment of genetic diversity among different biotypes of Physalis minima. Indian J. Weed Sci., 2017; 48(4): 417-420.
CrossRef

This work is licensed under a Creative Commons Attribution 4.0 International License.





