Manuscript accepted on : 13-01-2021
Published online on: 22-01-2021
Plagiarism Check: Yes
Reviewed by: Dr. Tsvetelina Velikova![]()
Second Review by: Dr. Oguzhan Özdemir
Final Approval by: Dr. Haseeb Ahmad Khan![]()

Gut Microbiome of Two Different Honeybee Workers Subspecies In Saudi Arabia.
Marfat Alatawy1,2* , Sanaa G. Al-Attas1
, Sanaa G. Al-Attas1 , Ahmad I. Assagaf1
, Ahmad I. Assagaf1 , Rashad R. Al-Hindi1
, Rashad R. Al-Hindi1 , Khalid M. Alghamdi1
, Khalid M. Alghamdi1 ,Jazem A. Mahyoub1
,Jazem A. Mahyoub1 , Alshehri D1,2
, Alshehri D1,2 , Al-Amrah H1
, Al-Amrah H1 , Alatawi H1,2
, Alatawi H1,2 , Edris S 1,3,4
, Edris S 1,3,4 , Ahmed Bahieldin1,3
, Ahmed Bahieldin1,3
1Department of Biological Sciences, Faculty of Science, King Abdulaziz University, Jeddah, Saudi Arabia.71491
2Department of Biology, College of Science, Tabuk University, Tabuk, Saudi Arabia.74191
3Department of Genetics, Faculty of Agriculture, Ain Shams University, Cairo, Egypt. 11241
4Princess Al Jawhara Albrahim Centre of Excellence in Research of Hereditary Disorders (PACER-HD), King Abdulaziz University, Jeddah, Saudi Arabia.
Corresponding Author E-mail: mevalatawi@ut.edu.sa
DOI : http://dx.doi.org/10.13005/bbra/2870
ABSTRACT:
Honeybees play a vital role in the world’s food supply by acting as essential pollinators in the agricultural fields. Interestingly, more than one third of the world’s essential crops are honeybee’s dependant. The adult honeybeeworkers harbour a simple specific bacterial spectrum in their guts with vital role in bees’ health. Gut microbial diversity of adult honeybee workerswasstudied through targeting the V3 and V4 regions of the 16S rRNA geneviaIllumina MiSeq. The study identified four phyla of the gut microbiomesinadult workersof the two-honeybee subspecies A.m. jemeniticaandA.m. carnica. The most abundant phylum in microbiome of A.m. jemeniticawasFirmicutes (48%), while Protobacteria and Actinobacteriaphylawere less abundantat figures of31% and 10%, respectively. In microbiome of A.m. carnica,Firmicutes (57%) was also the most dominant phylum, while Protobacteria and Actinobacteria had lower prevalence at figures of 31% and 10%, respectively. At genus level, adult honeybee workers harboured a number ofLactobacillus spp.in their guts with relative abundance of 80% in A.m. jemeniticaworkers compared to52%forA.m. carnicaworkers.Up toour knowledge, this is the first study of its kind on gut microbiome diversity inhoneybee workersof different origins conducted in Saudi Arabia using high-throughput 16S rRNA gene sequencing technology. The results indicatedthat the variability inmonophyletic origin of host of honeybee workers affectedgut microbiota composition.
KEYWORDS: A.M. Jemenitica; A.M. Carnica,16S Rrna; Diversity; High-Throughput Sequencing
Download this article as:| Copy the following to cite this article: Alatawy M, Al-Attas S. G, Assagaf A. I, Al-Hindi R. R, Alghamdi K. M, Mahyoub J. A, Alshehri D, Al-Amrah H, Alatawi H, Edris S, Bahieldin A. Gut Microbiome of Two Different Honeybee Workers Subspecies In Saudi Arabia. Biosci Biotech Res Asia 2020;17(4). |
| Copy the following to cite this URL: Alatawy M, Al-Attas S. G, Assagaf A. I, Al-Hindi R. R, Alghamdi K. M, Mahyoub J. A, Alshehri D, Al-Amrah H, Alatawi H, Edris S, Bahieldin A. Gut Microbiome of Two Different Honeybee Workers Subspecies In Saudi Arabia. Biosci Biotech Res Asia 2020;17(4). Available from: https://bit.ly/2MudhTg |
Introduction
Honeybees belong to the genus Apis, which is known for its tremendous role in pollination. Unfortunately, honeybee population is recently declining with a potential risk on the agricultural service and subsequently the food supply, not only locally in Saudi but also globally1. There is a known mutually beneficial relationship between honeybee gut microbiome and its host. The host provides the optimum environment for bacterial growth, while the bacterial community in honeybee guts aids in efficacy of nutrients absorption, optimum growth and development of the host and its ability to defend pathogens, and its adaptation to surrounding environment2.Honeybee gut represents a simple model system to study the relationshipbetween gut microbiome with honeybeehosts3,4.The bacterial community in adult honeybee workers is diverse and estimated to reach one billion bacterial cells in each worker’s gut5,6. Such a diversity in bacterial community is dependent on the type of flower that hosts the insect, as well as many other environmental factors7.Gut microbiome of honeybee (Apis mellifera) workers is composed ofeight to nine core species8,9, e.g., Bartonella apis10, Acetobacteraceae11,Parasaccharibacter11, Snodgrassella alvi12, Bifidobacterium asteroids13, Lactobacillus sp.14, Frischella perrara15and Gilliamella apicola12.
The two most common bee species that are widely distributed throughout the kingdom of Saudi Arabia are the indigenous Apis mellifera jementica, which is a native species, and Apis mellifera carnica, which is imported from Egypt16 as honey production of domestic bees does not meet the growing demands in Saudi Arabia. Moreover, the production cost is relatively high. Exotic bee colonies have been imported over time, reaching 200,000 bee packages annually16. It is well known for local beekeepers that the indigenous bees A.m. jementicahighly tolerates local stressful conditions when compared with exogenous races A.m. carnica, particularly during summer when the air temperature becomes extremely high. It is also noticed that at high temperatures, indigenous bees continue to forage for pollen and collect nectar, whereas imported bees will stop foraging16. Initial reports revealed that the subspecies of exotic honeybees have lower heat tolerance, shorter foraging durations and are more susceptible to Varroa mites when compared with indigenous bees16.
In the present study, we compered the gut microbiomecomposition and diversity of the adult honeybees of Apis mellifera jementica and Apis mellifera carnica in Saudi Arabia using high-throughput 16S rRNA gene sequencing technology.
Material and Methods
Sample collection,isolation of guts microbiota and DNA extraction
Five samples each from honeybee workers of A.m. jemenitica and A.m. carnicawere collected in November 2019 from a single hive of Beekeeper Cooperative Association at Al Baha, Saudi Arabia. The collected samples were immediately stored at −80°C.Forwhole gut dissection of honeybee workers,surface disinfection was done using 1 ml aqueous ethanol (70%, v/v) for 45 sec. Dissected guts were, then,placed in a pre-frozen mortar and 700μl S1 lysis buffer (Invitrogen, Thermo Fischer Scientific, USA) were added and guts were transferred to bead tube for extraction process.DNA of gut samples was extracted by the genomic DNA extraction kit (Invitrogen, Thermo Fischer Scientific, USA), and stored at -20°C for further molecular analysis.
PCR amplification
PCR was run to amplify bacterial 16S rRNA gene of the variable regions V3-V4. The two universal primers used for PCR are 341F 5′-ACTCCTACGGGAGGCAGCAG-3′ (forward primer) and 806R 5′- GGACTACHVGGGTWTCTAAT-3′ (reveres primer). The PCR conditions were set as the following: one cycle for initial denaturation at 95°C for 5 min; 25 cycles of denaturation at 95°C for 30sec followed by annealing at 56°C for 30 sec andprimer extension at 72°C for 40 sec; and a one cycle for final extension at 72°C for 10 min. The generated PCR products were checked for quality and selected products were utilized in preparing Illumina DNA libraries. DNA sequencing was run using Illumina Miseq platform (Illumina, San Diego, CA) at Beijing Genome Institute (BGI), China to generate high-quality pair-ends of ~300 bp.
Statistical analysis
The high quality paired reads produced in fasta files as raw data were de-multiplexed, quality-filtered and trimmed by trimmomatic package (Version 0.33) through Quantitative Insights Into Microbial Ecology 2 pipeline (QIIME2, v1.80). Obtained reads were merged into single sequence files by the Fast Length Adjustment of SHort reads (FLASH, Version 1.2.11). In order to assign generated unique sequences into operational taxonomic units (OTUs), reads were tagged and clustered into OTUs with similarity cut off of 97% using the de novo OTU piking procedure. Usearch (Version 7.0.1090)19 was, then, used to remove Chimeric sequences. Taxonomies were plotted against the gut Microbiome Database (HOMD RefSeq, Version 13.2) through the RDP classifier (Version 2.2)17 and the Green-genes database (version 2013051816S rDNA database, http://qiime.org/home_static/dataFiles.html) with a cut off of 70%. Alpha diversity indeceswere measured in order to assess the intra-species variations within a given sample using Mothur (v1.31.2). Alpha diversity and rarefaction curve boxplots were constructed using software R (v3.1.1). To investigate the inter-species variations within samples, the beta diversity matrices were conducted and visualized using principal coordinate analysis (PCoA) by package ‘ade4’ of software R (v3.1.1). Also, heat maps were generated using the package ‘gplots’ of software R (v3.1.1), and, then, sequence alignments were searched against the Silva core set (Silva_108_core_aligned_seqs) by using PyNAST ‘align_seqs.py’. The obtained OTU phylogenetic tree was, then, plotted by software R (v3.1.1), and visualized through QIIME2 (v1.80).
Annotation of generated OTUs was done in order to detect the relative abundance at different taxonomical levels (phylum, genus and species). Finally, Metastats, PERMANOVA and Benjamini–Hochberg false discovery rate (FDR) correction were also used to correct for multiple hypothesis. The Linear Discriminant Analysis (LDA) Effect Size (LEfSe) was applied using software LEfSewith the online interface Galaxy (version 1.0.0; http://huttenhower.sph.harvard.edu/galaxy/root),to discriminate the two taxonomic races determining highly presented bacterial taxon within each race depending on statistical significance.
Results
Statistics of 16S rRNA Sequence data
The five gut microbiome samples of A.m. carnicawere identified asC1 to C5, while the five gut microbiome samples of A.m. jemeniticawereidentified as J1 to J5. Illumina MiSeq was used in sequencing thepartial 16S rRNA gene and statistics details are tabulated in Table 1. A total of 2,279,519 sequence reads were obtained with an average length of 297 bp across different samples ranging from 293 to 300 bp. The average clean reads per subject were 218,741 and 237,162 for A.m. carnicaand A.m. jemeniticaworkers, respectively. A total of 2,269,665 tag-linked sequences were obtained across samples from both taxonomic groups with an average read of 236,125 and 217,807 per subject, for both A.m. carnicaand A.m. jemeniticaworkers, respectively (Figure S1). Furthermore, a total of 601,485 tags were obtained across samples with an average reads of 63,313 and 56,983 per subject, in both A.m. carnicaand A.m. jemeniticaworkers,respectively (Figure S1). The tagged sequences were assigned to a total of 171 OTUsacross samples ranging from 45 (J3) to 154 (C5) OTUs (Table S1). The sum of OTUs in A.m. carnica (A.M.C) is 373 with an average number of 74 OTUs, while 241 in A.m. jemenitica(A.M.J)with an average number of 48 OTUs.
Table 1: Statistics of deep sequencing data generated for gut microbiomes of 10 adult honeybee workers from subspecies A.m.carnica(C1-C5) andA.m.jemenitica (J1-J5).
| Sample ID | Reads length (bp) | Raw Data (Mbp) | N Base (%) | Low quality (%) | Clean Data (Mbp) | Data utilization (%) | Raw reads | Clean reads | Read utilization (%) |
| C1 | 300:300 | 157.62 | 0.035 | 3.863 | 143.01 | 90.73 | 2626982 | 2434712 | 92.68 |
| C2 | 299:300 | 160.13 | 0.028 | 4.042 | 144.97 | 90.53 | 2673272 | 2472472 | 92.49 |
| C3 | 298:300 | 145.38 | 0.024 | 4.128 | 131.32 | 90.33 | 2431182 | 2244372 | 92.32 |
| C4 | 297:300 | 154.8 | 0.022 | 4.032 | 140.04 | 90.47 | 2592892 | 2397372 | 92.46 |
| C5 | 296:300 | 90.52 | 0.025 | 4.629 | 80.83 | 89.29 | 1518842 | 1388132 | 91.39 |
| J1 | 297:293 | 149.45 | 0.021 | 3.73 | 136.59 | 91.4 | 2533052 | 2356712 | 93.04 |
| J2 | 296:293 | 162.1 | 0.026 | 3.963 | 147.33 | 90.89 | 2752092 | 2547162 | 92.55 |
| J3 | 294:293 | 145.85 | 0.02 | 3.664 | 133.58 | 91.59 | 2484682 | 2316072 | 93.21 |
| J4 | 293:293 | 135.37 | 0.021 | 3.797 | 123.52 | 91.25 | 2310112 | 2148432 | 93 |
| J5 | 300:293 | 158.64 | 0.032 | 3.702 | 145.15 | 91.5 | 2675232 | 2489772 | 93.07 |
Alpha diversity and principle coordinate analyses and rarefaction curve measurement
Alpha diversity indices were used to analyse the complexity of the included species. These indices are observed species (Sobs), Chao1, Ace, Shannon and Simpson. The Sobs and Chao1 indices indicated significance differences between A.M.C and A.M.Jgroups with higher diversity in A.M.C group. The P-values determined were 0.03175 for Sob and0.01587 for Chao1 indices (Figure 1 and Table 2). On theother hand, Shannon and Simpson indices revealed no significant difference between A.M.C and A.M.Jgroups. Chao1 and Shannon indicesreflect the species diversity in terms of richness, while Simpson index isindicative of evenness20.The Simpson values in A.M.C and A.M.J groups were 0.1377 and 0.12313, respectively, while, Shannon index values were 2.4658 and 2.4769 in A.M.C and A.M.J groups, respectively(Figure 1 and Table 2).
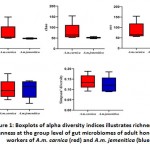 |
Figure 1: Boxplots of alpha diversity indices illustrates richness and evenness at the group level of gut microbiomes of adult honeybee workers of A.m. carnica (red) and A.m. jemenitica (blue) |
Table 2: Alpha diversity comparison results among groups of gut microbiomes of honeybee workers from subspeciesA.m.carnica(A.M.C) andA.m.jemenitica (A.M.J).
| Alpha diversity measure | Mean (A.M.C) | SD (A.M.C) | Mean (A.M.J) | SD (A.M.J) | p-value |
| Sobs | 74.6 | 44.59036 | 48.2 | 3.11448 | 0.03175 |
| Chao | 79.1 | 42.37209 | 52.01667 | 3.76128 | 0.05556 |
| Ace | 83.69118 | 40.78638 | 53.23999 | 3.78296 | 0.01587 |
| Shannon | 2.4658 | 0.22593 | 2.4769 | 0.21404 | 0.84127 |
| Simpson | 0.1377 | 0.0347 | 0.12313 | 0.0389 | 0.30952 |
Principal coordinate analysis (PCoA) was used to display the diversity as well as the differences in OTU composition.Diversity of A.M.C subjects was higher towards positive and negative PCA 1 directions (PC1), whereasthat of A.M.J subjects was higher towards positive and negative PCA 2 directions (PC2). As an overall picture, the diagram shows that the mean value of A.M.C group was localized in positive portion of PC1 and negative portion of PC2, whereasA.M.J group was mainly localized in the positive portion of PC2(Figure 2). The principal coordinate analysis (PCoA) plots were created using a Bray-Curtis distance matrix and the samples were plotted to represent the microbial community compositional differences between samples. The plots are dimensionally scattered in accordance to their gut microbiome compositional relationships. The results of the present study indicate that the differences ingut microbiomes between these two groups arepossibly due to the different origins of worker honeybeesof the two subspecies.
The stacked number of OTUs and the number of observed species for different samples as rarefaction measures are shown in Figure S2. When the refraction curves inclines (Figure S2a) or stops climbing (Figure S2b), the produced data would be enough for further analysis. However, as long as the curve is still climbing, the complexity of the data in samples become higher; since more species being detected throughout sequencing analysis. The two rarefaction curve measures refer to the maximum number of sequences attained for all samples that allows to study taxonomic relative abundance and to assess eligibility of such data to represent all species of any microbial community. The findings from both rarefaction measures show that 54,000 is the maximumnumber of sequence reads that can be used further instudyingtaxonomic abundance (Figure S2).
Structure of gut microbiomes across the two honeybee workers
Two taxonomic ranks (phylum and species) were used in the comparison of gut microbiomesbetween adult honeybee workers A.M.CandA.M.J at the phylogenetic level (Figure 3). The results indicate that phylumFirmicutesharbours 24 genera,while Proteobacteria,Actinobacteria, Bacteroidetes andThermiharbour 23, 8, 6 and 2 genera, respectively (Figure 3).
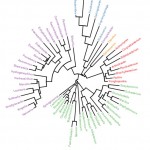 |
Figure 3: Phylogenetic tree at genus level of gut microbiomes of adult honeybee workers of A.m.carnica and A.m.jemenitica. Genera having the same color belong to the same phylum. |
Differential abundance of microbes due to different origin of worker
The observed microbial taxa along with their redundancies across different samples identified after OTU annotation are described in Table S2. The taxa refer to phylum, class, order, family, genus, and species. Eight phyla of the gut bacteria were identified according to relative abundance. They are Actinobacteria, Bacteroidetes, Cyanobacteria, Firmicutes, Protobacteria, TM7, Tenericutes and Thermi(Figure 4). Aligning with the number of genera of each phylum shown in Figure 3, the most abundant phylum were Firmicutes (57%), Protobacteria (31%) and Actinobacteria (10%) inA.M.C group (Figure 4).
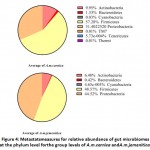 |
Figure 4: Metastatsmeasures for relative abundance of gut microbiomes at the phylum level forthe group levels of A.m.carnica andA.m.jemenitica. |
Meanwhile, Firmicutes (48%), Protobacteria (44%) and Actinobacteria (6%) were the most abundant in A.M.J group (Figure 4). The comparison at phylum level revealed a significant increase in Cyanobacteria in the A.M.C group (P-value = 0.031746), while a significant increaseof Protobacteria in the A.M.J group (P-value = 0.037724)(Table S3). Interestingly, Table S3 also indicates the existence of the three phyla TM7, Tenericutes and Thermi only in A.M.C group. The previous results align with those ofthe heat map at phylum level asFirmicutes, Protobacteria and Actinobacteria were shown to be the most abundant phyla across samples and groups (Figure S3).
In terms of species relative abundance in the gut microbiomes of two groups A.M.C and A.M.J Bacteroides_fragilis, Bacteroides_ovatus, Commensalibacter_intestini, Blautia_producta, Melissococcus_plutonius, Ruminococcus_gnavus,Saccharibacter_floricola and Snodgrassella_alviwere shown to be the most abundant (Figure 5).
The figure also indicates that a large proportion of the OTUs were not assigned to a certain species (93.80% for A.M.C and 86.20% for A.M.J). We have no explanation for these results except that a large number of species in workers of honeybee was not identified or classified before. The results in Table S4 indicates asignificant increase of Melissococcus_plutoniusin the gut microbiome of A.M.C (P-value = 0.034454), while Snodgrassella_alvi in theA.M.J group (P-value =0.008948). Results for the latter species Snodgrassella_alvi align with that presented in Figure 5c. TheRuminococcus_gnavusandSaccharibacter_floricolawere not existed in theA.M.J group. The heat map at species level indicates that Snodgrassella_alviharbours the highest relative abundanceacross all samples (Figures S4).
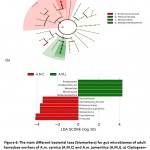
Linear discriminant analysis effect size (LEfSe) and its LDA scores (˃ 3) wereused to identify possible biomarkers in gut microbiota that refer to the origin of the host (Figure 6). The results in cladogram indicate that the possible marker in gut microbiome of A.M.C is Enterococcaceae family, while Neisseriaceaea, Neisseriales and Betaproteobacteria taxa of A.M.J (Figure 6a). Biomarkers in A.M.C based on LDA score includeEnterococcaceae, Saccharibactersp.,Saccharibacterflorcola, Firmicutes (Melissococcussp. andMelissococcusplutonius) and Cyanobacteria while Betaproteobacteria (Neisseriales, Neisseriaceae,Snodgrassella sp. andSnodgrassella_alvi)in A.M.J (Figure 6b).
Discussion
The gut microbiomestructure of honeybee workersis dependent upon monophyletic origin of the host9, social interactions8and the type of diet consumed, whether workers are beebread, pollen or nectar1.In the present study, high-throughput sequencing was carried out for samples taken from the two honeybee subspecies A.m. carnicaandA.m. jemenitica and statistical analysis proved that the diversity of the bacterial community composition ofA.m. carnicaandA.m. jemenitica was statistically significant.
Four major bacterial phyla (Firmicutes, Proteobacteria, Actinobacteria and Bacteroidetes) were recognized in the guts of honeybee workers of the two subspecies A.m. carnicaand A.m. jemenitica. The dominant phylum ingut microbiomes of the two subspecies wasFirmicutes with values of 57.2 and 48.5%, respectively. This conclusion was also drawn in several previous reports9,21,22,23.Genus Lactobacillus, gram-positive bacteria belongingto the family Lactobacillaceae (Firmicutes), was found to have a high relative abundance in adult workers of both A.m. carnicaandA.m. jemeniticawith values of 52% and 80%, respectively. It is a core gut bacterium that is dominant in the rectum of honeybee workers. Within this context, Ahnet al.24 concluded thatLactobacillaceaedominates in both of A. cerana and A. mellifera species.This genus produces several compounds in honeybee gut with known antimicrobial activities such as organic acids, hydrogen peroxide, bacteriocin, reutericyclin and reuterin that mostly inhibit decaying and protects againstpathogenic bacteria, as well as some fungi25,26.Therefore,Honeybeeslikely use lactobacilli as probiotic27.In the present study, the dominance of Lactobacilli in both A.m. carnicaandA.m. jemeniticaadult workers is supported by the presence of low pH (3.9) of honey and nectar28. This is concluded because of the ability of lactobacilli to ferment sugar in the gut of honeybee workers and, hence,to generate acidic environment29, which inhibits the growth of many other bacteria. The low abundance in Lactobacillaceaewas reported to be associated with the presence of pathogenic bacteria30.
GenusBifidobacterium,gram-positive bacteria belonging to the Actinobacteria phylum, was also identified in gut of both A.m. carnicaandA.m. jemenitica adult workers. Again, it is dominant in rectum, and a core gut bacteria of honeybee workers.Bifidobacterium strains carry large surface proteins, which have a role in adhesion or degradation of plant materials7,31,32. Additionally, Bifidobacteriumcarriesgene clusters that are responsible for the production and utilization of trehalose, which is a disaccharide molecule used by insects as an energy reservoir, in comparison to glycogen, which is the energy storage form in mammals33.
Family Neisseriaceaeand its descendentSnodgrassella_alvi(S. alvi), gram-negative bacteria belonging toBetaproteobacteria phylum, significantly increased in A.m. jemenitica.These bacteria participate in oxidation of carbohydrates. However, the pathway for the uptake and glycolytic breakdown of carbohydrates does not exist in S. alvi, thus,this bacteriumis located consistently within the periphery of the insect’s gut lumen. This area has high oxygen concentrations and this environment is preferable for S. alvidue to its dependence on aerobic respiration34,35. Insects depend on the aerobic oxidation of carboxylates rather than breaking down carbohydrates resulting in various products such as citrate, malate, acetate and lactic acid that serve as energy sources12,27. The steady co-exits of S. alvi with other fermentative bacterial taxa in the same gastrointestinal environment can result from utilizing separate sets of resources leading to metabolic variations suggesting a syntrophic interaction. For example, S. alvican utilize some of the substrates such as lactic acid, acetate and formate, which are produced from carbohydrate fermentation36,37. Furthermore, S. alvi and G. apicola38are enriched with genes encoding biofilm formation. The two species inhabit the host’s ileum, indicating that the biofilm can provide a protective layer against pathogens.
The bacteria of the family Acetobacteraceaeand its descendent genusCommensalibacter(also referred to as Alpha 2.1), gram-negative bacteria belonging to phylum Proteobacteria,wereidentifiedas a core member of the gut microbiota in honeybees and bumble bees9,31. It was observed mainly in the midgut and hindgut of honeybee workers. In our study, Commensalibacterpresents in A.m. carnicaandA.m. jemenitica. However, Saccharibacterflorica (Alpha-2.2) presents only in A.m. carnica. Furthermore, Saccharibacterflorica is isolated from pollen, suggesting that this phylotype is associated with flowers39.The role of these phylotypes (Alpha 2.1 andAlpha-2.2) is associatedwith their abilities to adaptwith fast growing metabolic processes, with two distinctive mechanisms. Alpha2.1 bacteria harvest energy througha wide range of substrates linkedand utilizedthrough a flexible oxidative and biosynthetic metabolism.Whereas, Alpha2.2 bacteria, that lack alternative oxidative pathways, determine metabolic processes through oxidative fermentation after harvesting glucose for rapid energy40.
The bacteria of the familyEnterococccaceaeand its descendent species Melissococcusplutonius, gram-positive bacteria of phylum Firmicutes, present in low abundance (3%) in gut microbiome ofA.m. carnica honeybee workers. This conclusion was also noted in previous reports41.M. plutoniusis known to cause the European foulbrood (EFB) in earlystage of honeybee larvae, with assistance from secondary invaders (Enterococcus faecalis, Paenibacillus alvei and Bacillus pumilus). M. plutonius wasshown to have 30 different sequence types clustered under three clonal complexes (CC 3, CC12, and CC13)42,44, where CC13is the least virulent complex43,45. Honeybee workers transmit M. plutonius between colonies via robbing and drifting46,47.Erban et al.45 compared control samples from the EFB zone with samples from EFB zone without clinical symptoms,and bees from colonies from EFB zone with clinical symptoms. The study identified a 100-fold higher prevalence of M. plutonius in colonies with EFB symptoms, while it only presents in 3 of 16 control colonies that are distant from the EFB zone. This suggests that M. plutonius has lower abundance in healthy honeybee colonies, which is consistent with the results of the present study.
Conclusion
The present findings indicative that differences in gut microbiome structuresof honeybee workers of the two subspecies A.m. carnicaand A.m. jemeniticaare due to varied monophyletic origin of the host. These findings support previous results suggesting that honeybee workers have a mutual coevolving relationship with specific group of bacteria. This group of bacteria co-exists and is maintained throughout the descending generations of the host.Inclusion of more subspecies inhabited in Saudi Arabia along with ones of this study can further support our findings.
Supplementary Information
Additional file: Additional Table S1, S3 and S4 Additional Figure S1 to S4
Additional file: Additional Table S2
Acknowledgement
This study was supported by Beekeeper Cooperative Association-Al Baha. The authors would like to thank Prof. Dr.Ahmad Al-Ghamdi, Dept. Plant Protection, College of Food and Agricultural Sciences, KSU, Riyadh, for his support and cooperation during this study.
Conflict of Interest
The authors declare no conflict of interest.
References
- Powell, J. E., Martinson, V. G., Urban-Mead, K. & Moran, N. A. Routes of acquisition of the gut microbiota of the honeybeeApis mellifera. Environ. Microbiol.80, 7378–7387 (2014).
CrossRef - Jones, J. C. et al. Gut microbiota composition is associated with environmental landscape in honeybees. Ecology and evolution8, 441–451 (2018).
CrossRef - Engel, P. & Moran, N. A. The gut microbiota of insects–diversity in structure and function. FEMS microbiology reviews37, 699–735 (2013).
CrossRef - Krishnan, M. et al. Insect gut microbiome–An unexploited reserve for biotechnological application. Asian Pacific journal of tropical biomedicine4, S16–S21 (2014).
CrossRef - Martinson, V. G., Moy, J. & Moran, N. A. Establishment of characteristic gut bacteria during development of the honeybee worker. Environ. Microbiol.78, 2830–2840 (2012).
CrossRef - Moran, N. A. Genomics of the honeybee microbiome. Current opinion in insect science10, 22–28 (2015).
CrossRef - Jones, J. C. et al. The gut microbiome is associated with behavioural task in honeybees. Insectessociaux65, 419–429 (2018).
CrossRef - Kapheim, K. M. et al. Caste-specific differences in hindgut microbial communities of honeybees (Apis mellifera). PloS one10, e0123911 (2015).
CrossRef - Martinson, V. G. et al. A simple and distinctive microbiota associated with honeybees and bumble bees. Molecular Ecology20, 619–628 (2011).
CrossRef - Kešnerová, L., Moritz, R. & Engel, P. Bartonella apis sp. nov., a honeybee gut symbiont of the class Alphaproteobacteria. International journal of systematic and evolutionary microbiology66, 414–421 (2016).
CrossRef - Corby-Harris, V. et al. Origin and Effect of Alpha 2.2 Acetobacteraceae in Honeybee Larvae and Description of Parasaccharibacterapium gen. nov., sp. nov. Environ. Microbiol.80, 7460–7472 (2014).
CrossRef - Kwong, W. K. & Moran, N. A. Cultivation and characterization of the gut symbionts of honeybees and bumble bees: description of Snodgrassellaalvi gen. nov., sp. nov., a member of the family Neisseriaceae of the Betaproteobacteria, and Gilliamellaapicola gen. nov., sp. nov., a memb. International journal of systematic and evolutionary microbiology63, 2008–2018 (2013).
CrossRef - Bottacini, F. et al. Bifidobacterium asteroides PRL2011 genome analysis reveals clues for colonization of the insect gut. PLoS One7, e44229 (2012).
CrossRef - Olofsson, T. C., Alsterfjord, M., Nilson, B., Butler, È. & Vásquez, A. Lactobacillus apinorum sp. nov., Lactobacillus mellifer sp. nov., Lactobacillus mellis sp. nov., Lactobacillus melliventris sp. nov., Lactobacillus kimbladii sp. nov., Lactobacillus helsingborgensis sp. nov. and Lactobacillus kullabergensis sp. nov., isol. International journal of systematic and evolutionary microbiology64, 3109 (2014).
CrossRef - Engel, P., Kwong, W. K. & Moran, N. A. Frischellaperrara gen. nov., sp. nov., a gammaproteobacterium isolated from the gut of the honeybee, Apis mellifera. International journal of systematic and evolutionary microbiology63, 3646–3651 (2013).
CrossRef - Zhu, X., Wang, J., Reyes-Gibby, C. &Shete, S. Processing and Analyzing Human Microbiome Data. in Statistical Human Genetics 649–677 (Springer, 2017).
CrossRef - Cole, J. R. et al. Ribosomal Database Project: data and tools for high throughput rRNA analysis. Nucleic acids research42, D633–D642 (2014).
CrossRef - DeSantis, T. Z. et al.Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Applied and environmental microbiology72, 5069–5072 (2006).
CrossRef - Edgar, R. C. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nature methods10, 996–998 (2013).
CrossRef - Lombogia, C. A., Tulung, M., Posangi, J. &Tallei, T. E. Bacterial Composition, Community Structure, and Diversity in Apisnigrocincta Gut. International Journal of Microbiology2020, (2020).
CrossRef - Jeyaprakash, A., Hoy, M. A. & Allsopp, M. H. Bacterial diversity in worker adults of Apis mellifera capensis and Apis mellifera scutellata (Insecta: Hymenoptera) assessed using 16S rRNA sequences. Journal of invertebrate pathology84, 96–103 (2003).
CrossRef - Mohr, K. I. &Tebbe, C. C. Diversity and phylotype consistency of bacteria in the guts of three bee species (Apoidea) at an oilseed rape field. Environmental Microbiology8, 258–272 (2006).
CrossRef - Yoshiyama, M. & Kimura, K. Bacteria in the gut of Japanese honeybee, Apiscerana japonica, and their antagonistic effect against Paenibacillus larvae, the causal agent of American foulbrood. Journal of Invertebrate Pathology102, 91–96 (2009).
CrossRef - Ahn, J.-H. et al. Pyrosequencing analysis of the bacterial communities in the guts of honeybees Apiscerana and Apis mellifera in Korea. Journal of Microbiology50, 735–745 (2012).
CrossRef - Lahtinen, S., Ouwehand, A. C., Salminen, S. & von Wright, A. Lactic acid bacteria: microbiological and functional aspects. (Crc Press, 2011).
CrossRef - Mokoena, M. P. Lactic acid bacteria and their bacteriocins: classification, biosynthesis and applications against uropathogens: a mini-review. Molecules22, 1255 (2017).
CrossRef - Kwong, W. K., Mancenido, A. L. & Moran, N. A. Genome sequences of Lactobacillus sp. strains wkB8 and wkB10, members of the Firm-5 clade, from honeybee guts. Genome Announc.2, e01176-14 (2014).
CrossRef - JA, S. Cliver DO. Microorganisms in honey. J. Food Microbiol31, 1–26 (1996).
CrossRef - Bignell, D. E. & Heath, L. A. F. Electropositive redox state of the fifth-instar larval gut of Apis mellifera. Journal of apicultural research24, 211–213 (1985).
CrossRef - Koch, H. & Schmid-Hempel, P. Socially transmitted gut microbiota protect bumble bees against an intestinal parasite. Proceedings of the National Academy of Sciences108, 19288–19292 (2011).
CrossRef - Kwong, W. K. & Moran, N. A. Gut microbial communities of social bees. Nature Reviews Microbiology14, 374–384 (2016).
CrossRef - Bonilla-Rosso, G. & Engel, P. Functional roles and metabolic niches in the honeybee gut microbiota. Current opinion in microbiology43, 69–76 (2018).
CrossRef - Alatawy, M. et al. Gut Microbial Communities of Adult Honeybee Workers (Apis Mellifera). Biosciences Biotechnology Research Asia17, (2020).
CrossRef - Brune, A. Symbiotic digestion of lignocellulose in termite guts. Nature Reviews Microbiology12, 168 (2014).
CrossRef - Egert, M. et al. Structure and topology of microbial communities in the major gut compartments of Melolonthamelolontha larvae (Coleoptera: Scarabaeidae). Environ. Microbiol.71, 4556–4566 (2005).
CrossRef - Anderson, K. E. et al. Hive‐stored pollen of honeybees: many lines of evidence are consistent with pollen preservation, not nutrient conversion. Molecular ecology23, 5904–5917 (2014).
CrossRef - Corby-Harris, V., Maes, P. & Anderson, K. E. The bacterial communities associated with honeybee (Apis mellifera) foragers. PloS one9, e95056 (2014).
CrossRef - Engel, P., Martinson, V. G. & Moran, N. A. Functional diversity within the simple gut microbiota of the honeybee. Proceedings of the National Academy of Sciences109, 11002–11007 (2012).
CrossRef - Jojima, Y. et al.Saccharibacterfloricola gen. nov., sp. nov., a novel osmophilic acetic acid bacterium isolated from pollen. International Journal of Systematic and Evolutionary Microbiology54, 2263–2267 (2004).
CrossRef - Bonilla-Rosso, G. et al.Acetobacteraceae in the honeybee gut comprise two distant clades with diverging metabolism and ecological niches. bioRxiv 861260 (2019).
CrossRef - Sopko, B. et al. Detection and quantification of Melissococcusplutonius in honeybee workers exposed to European foulbrood in Czechia through conventional PCR, qPCR, and barcode sequencing. Journal of Apicultural Research 1–12 (2019).
- Budge, G. E. et al. The occurrence of Melissococcusplutonius in healthy colonies of Apis mellifera and the efficacy of European foulbrood control measures. Journal of invertebrate pathology105, 164–170 (2010).
CrossRef - Budge, G. E. et al. Molecular epidemiology and population structure of the honeybee brood pathogen Melissococcusplutonius. The ISME journal8, 1588–1597 (2014).
CrossRef - Takamatsu, D. et al. Typing of Melissococcusplutonius isolated from European and Japanese honeybees suggests spread of sequence types across borders and between different Apis species. Veterinary microbiology171, 221–226 (2014).
CrossRef - Erban, T. et al. European foulbrood in Czechia after 40 years: application of next-generation sequencing to analyzeMelissococcusplutonius transmission and influence on the bacteriome of Apis mellifera. PeerJ Preprints4, e2618v1 (2017).
CrossRef - Hunt, M. G. Index to Department Bulletins. (US Government Printing Office, 1936).
- Belloy, L. et al. Spatial distribution of Melissococcusplutonius in adult honeybees collected from apiaries and colonies with and without symptoms of European foulbrood. Apidologie38, 136–140 (2007).
CrossRef

This work is licensed under a Creative Commons Attribution 4.0 International License.