Manuscript accepted on : 27/12/2019
Published online on: --
Plagiarism Check: Yes
Reviewed by: Muhammad, Naoshad
Second Review by: Eshak Bahbah
Final Approval by: Ashraf
Mohd Suhail1, Asma perveen1 , Amjad Husain1* and Mohd Rehan*2,3
, Amjad Husain1* and Mohd Rehan*2,3
1School of Life Sciences, The Glocal University, Saharanpur, Uttar Pradesh, 247121, India
2King Fahd Medical Research Center, King Abdulaziz University, Jeddah, Saudi Arabia
3Department of Medical Laboratory Technology, Faculty of Applied Medical Sciences, King Abdulaziz University, Jeddah, Saudi Arabia
Corresponding Author's Email: amjad@theglocaluniversity.in
DOI : http://dx.doi.org/10.13005/bbra/2787
ABSTRACT: Nuclear factor kappa B (NF-κB), a transcription factor is a well-established cancer therapeutic target. NF-κB’s linkage with cancer is known through the constitutive activation of NF-κB in several cancer types. The most important role of NF-κB as a transcription factor is its ability to promote cell survival through the induction of transcription of target pro-survival genes and thus inhibition of programmed cell death (PCD) by resulting proteins in both malignant and normal cells. Current findings have unveiled that green tea catechins exert anticancer effect by inhibiting the activity of various receptors including NF-κB. The current study is designed to gain the structural insights for inhibitory mechanism of catechin derivatives against NF-κB. The major green tea catechins include (-)-epicatechin (EC), (-)-epigallocatechin (EGC), (-)-epicatechin gallate (ECG), and (-)-epigallocatechin gallate (EGCG) and are included in the current study. The study explored the binding pose, interacting residues, molecular interactions, and predicted binding energy and dissociation constant for the catechin derivatives. Our results showed that the catechin derivatives bound well in the DNA binding site with adequate binding strength scores. The study suggested that the four catechin derivatives may act as potential inhibitors of NF-κB and thus, may inhibit the progression of various cancer types.
KEYWORDS: Nuclear Factor Kappa B; NF-κB; Catechin Derivatives; Epicatechin; EC, Epigallocatechin; EGC; Epicatechin-3-Gallate; ECG; Epigallocatechin-3-Gallate; EGCG and Cancer
Download this article as:| Copy the following to cite this article: Suhail M, Perveen A, Husain A, Rehan M. Exploring Inhibitory Mechanisms of Green Tea Catechins as Inhibitors of a Cancer Therapeutic Target, Nuclear Factor-κB (NF-κB) Biosci Biotech Res Asia 2019;16(4). |
| Copy the following to cite this URL: Suhail M, Perveen A, Husain A, Rehan M. Exploring Inhibitory Mechanisms of Green Tea Catechins as Inhibitors of a Cancer Therapeutic Target, Nuclear Factor-κB (NF-κB) Biosci Biotech Res Asia 2019;16(4). Available from: https://bit.ly/37Z4WNb |
Introduction
Cancer is one of the major chronic diseases worldwide, and every year 12.7 million new incidences are recorded. This number is expected to reach 26 million by 20301. The emerging targeted therapies provide a lot of hope, whereby a single target protein or enzyme is blocked using an inhibitor. The accumulating evidence established nuclear factor kappa B (NF-κB) as an excellent cancer therapeutic target2, 3. The NF-κB, which acts as a transcription factor, exists in the form of homo or heterodimeric complex. These complexes are made up of Rel-like domain including proteins RelA (p65), RelB, p105 (NFκB1), (p50) NFκB1, Rel and p52 (NFκB2), and the complex from two different subunits p65 and p50 seems to be the major one. NF-κB’s linkage with cancer is shown through the constitutive activation of NF-κB in several types of cancer like breast, lung, liver, pancreatic, prostate and various kinds of lymphoma4-9. The most important role of NF-κB is its ability to promote cell survival through the transcription of target pro-survival genes, resulting in protein expression that inhibits the programmed cell death (PCD) mechanism in both types of malignant and normal cells10, 11. Due to the broad significance of NF-κB, it has been challenging to target NF-κB in cancer cells12. Last few years’ studies have unveiled that NF-κB is activated by carcinogens, chemotherapeutic agents, tumour promoters, and by inflammatory cytokines13. Studies on preclinical models have shown that several chemotherapy drugs like taxanes, anthracyclines and platinum-based agents induce the activation of NF-κB pathway14, while there are a majority of drugs identified to be the inhibitor of NF-κB signaling, with potencies as low as 20nM. Some of these drugs like narasin, sunitinib malate, lestaurtinib, tribromsalan, bithionol, fluorosalan and emetine inhibit NF-κB signaling via inhibition of IkBa phosphorylation15.
The natural compounds have increasingly been exploited by the scientific community for anticancer potential16-19. In this view, green tea catechins were extensively studied for anticancer mechanisms controlling cell proliferation, cell death in tumour cells and vascular angiogenesis in solid tumours20, 21. Recent studies have suggested that natural compounds catechins found in green tea have inhibitory activity against the NF-κB22. There are four major components in green tea found as catechins; 1) (-)-epicatechin (EC), 2) (-)-epigallocatechin (EGC) 3) (-)-epicatechin-3-gallate (ECG) and 4) (-)-epigallocatechin-3-gallate (EGCG) (Fig.1). EGCG and EGC are the most abundant in green tea23, 24. Current findings have unveiled that green tea catechins particularly interact with cell surface receptors, including trans-membrane receptor 67L, lipid rafts, receptor tyrosine kinases (RTKs) and endoplasmic reticulum (ER)25. These catechins modulate gene expression through a direct effect on transcription factors or indirectly by epigenetic mechanism, and also interfere with intracellular proteostasis at various levels26. Recent developments in the advancement of knowledge in this field have shown that EGCG shows an anticancer effect by inhibiting the activity of NF-κB and several other receptors22, 27. In recent years, the computational studies are increasingly being used for exploring the inhibitory mechanism of active compounds and in drug designing28-34. Despite the knowledge of anticancer potential and NF-κB inhibitory activity, the exact binding poses and molecular interactions of these catechins for NF-κB were not explored yet. The current study is aimed to explore the inhibitory mechanism of catechin derivatives against NF-κB DNA binding activity through computational methods.
Materials and Methods
Data Retrieval
The 3-D structure of human transcription factor NF-κB p50 subunit in complex with DNA was retrieved from protein data bank with PDB ID, 1svc. The molecular structures of (-)-epicatechin (EC), (-)-epigallocatechin (EGC), (-)-epicatechin gallate (ECG), and (-)-epigallocatechin gallate (EGCG) were obtained from PubChem with compound IDs, CID: 72276, 72277, 107905, and 65064 respectively.
Molecular Docking
Dock v.6.535 was used for docking simulation of all four catechin derivatives into the DNA binding site of NF-κB. The binding site was considered as the residues around 5 Å vicinity of bound DNA. Chimera v.1.6.236 was used in initial structure preparation of protein and ligands.
Analyses of Docked Protein-Ligand Complex
Pymol v.1.337 was used to generate illustrations and to analyze the protein-ligand complexes. LigPlot v.1.4.3 program38 was used to generate protein-ligand interaction plots and to analyze the polar and non-bonding interactions between the protein and the ligands. X-Score v.1.2.1139 was used for binding energy and dissociation constant calculation prediction from the protein-ligand complex.
Result and Discussion
Molecular Docking of EC to NF-κB
Epicatechin (EC) was found to bound deep in the binding site of NF-κB and interacted with eight residues namely Tyr-60, Lys-147, Asp-209, Leu-210, Ser-211, Lys-244, Pro-246 and Asn-247 (Fig. 2). The EC forms 28 non-bonding interactions and two hydrogen bonds (through Leu-210 and Asn-247) with the binding site stabilizing the protein-ligand complex (Table 1). Of eight interacting residues, Lys-244 was proposed to be the key interacting residue as it was involved in maximum number (10) of non-bonding interactions. The high absolute values for the dock score (-32.01), binding energy (-7.14 kcal/mol), and dissociation constant (pKd, 5.23) also suggested towards quality binding of EC to NF-κB protein and therefore it may serve as a good NF-κB inhibitor for DNA binding activity (Table 2).
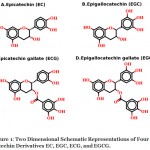 |
Figure 1: Two dimensional schematic representations of four catechin derivatives EC, EGC, ECG, and EGCG. |
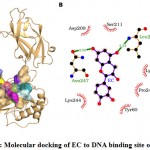 |
Figure 2: Molecular docking of EC to DNA binding site of NF-κB. |
Table 1: The transcription factor NF-κB residues interacting with EC are listed with the number of non-bonding contacts and hydrogen bonds.
| Interacting residues | Hydrogen bonds | Non-bonded contacts |
| Tyr-60 | 3 | |
| Lys-147 | 1 | |
| Asp-209 | 2 | |
| Leu-210 | 1 | 3 |
| Ser-211 | 4 | |
| Lys-244 | 10 | |
| Pro-246 | 2 | |
| Asn-247 | 1 | 3 |
Molecular Docking of EGC to NF-κB
The molecular docking results showed that EGC bound well to the NF-κB binding site and interacted with seven residues namely Tyr-60, Asp-209, Leu-210, Ser-211, Lys-244, Pro-246 and Asn-247 (Fig. 3). The seven interacting residues formed 24 non-bonding interactions and two hydrogen bonds (through Leu-210 and Asn-247) with EGC stabilizing the protein-ligand complex (Table 3). Of seven interacting residues, Lys-244 was proposed as the key interacting residue as it was involved in maximum (9) non-bonding interactions (Table 3). In addition to 24 non-bonding interactions and two hydrogen bonds, the high absolute values of dock score (-33.12), binding energy (-7.08 kcal/mol), and dissociation constant (pKd, 5.19) also indicated towards quality docking and thus EGC may also serve as potential inhibitor for DNA binding activity of NF-κB (Table 2).
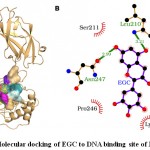 |
Figure 3: Molecular docking of EGC to DNA binding site of NF-κB. |
Table 2: The values for the binding strength scores (dock score, binding energy, and Kd, dissociation constants) are provided for the docked catechin derivatives.
| Compound | Dock score | Binding energy (kcal/mol) | pKd /-logKd |
| EC | -32.01 | -7.14 | 5.23 |
| EGC | -33.12 | -7.08 | 5.19 |
| ECG | -43.72 | -7.84 | 5.75 |
| EGCG | -42.70 | -7.78 | 5.70 |
Table 3: The transcription factor NF-κB residues interacting with EGC are listed with the number of non-bonding contacts and hydrogen bonds.
| Interacting residues | Hydrogen bonds | Non-bonded contacts |
| Tyr-60 | 3 | |
| Asp-209 | 2 | |
| Leu-210 | 1 | 3 |
| Ser-211 | 2 | |
| Lys-244 | 9 | |
| Pro-246 | 2 | |
| Asn-247 | 1 | 3 |
Molecular Docking of ECG to NF-κB
The molecular docking of ECG to NF-κB revealed that ECG bound well in the binding site and interacted with 10 residues including Tyr-60, His-144, Thr-146, Lys-147, Asp-209, Leu-210, Ser-211, Lys-244, Ala-245 and Pro-246 (Fig. 4). These 10 residues formed 46 non-bonding interactions and four hydrogen bonds making the protein-ligand complex stable (Table 4). Of 10 interacting residues, Lys-244 was found to be the key residue as it was involved in the maximum number (10) of non-bonding interactions and one hydrogen bond (Table 4). The absolute values of binding strength scores: dock score (-43.72), binding energy (-7.84 kcal/mol), and dissociation constant (pKd, 5.75) indicated towards quality binding and thus ECG may serve as a potential NF-κB inhibitor for DNA binding activity (Table 2).
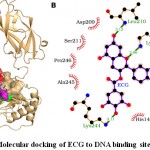 |
Figure 4: Molecular docking of ECG to DNA binding site of NF-κB. |
Table 4: The transcription factor NF-κB residues interacting with ECG are listed with the number of non-bonding contacts and hydrogen bonds.
| Interacting residues | Hydrogen bonds | Non-bonded contacts |
| Tyr-60 | 8 | |
| His-144 | 1 | |
| Thr-146 | 3 | |
| Lys-147 | 2 | 8 |
| Asp-209 | 5 | |
| Leu-210 | 1 | 2 |
| Ser-211 | 3 | |
| Lys-244 | 1 | 10 |
| Ala-245 | 3 | |
| Pro-246 | 3 |
Molecular Docking of EGCG to NF-κB
The molecular docking results showed that EGCG bound deep in the NF-κB binding site and formed interaction with 10 residues namely Tyr-60, His-144, Thr-146, Lys-147, Asp-209, Leu-210, Ser-211, Lys-244, Ala-245 and Pro-246 (Fig. 5). The ten interacting residues exerted 47 non-bonding interactions and three hydrogen bonds with EGCG and made the protein-ligand complex stable (Table 5). Of 10 interacting residues, Lys-244 was found to be the key residue as it was involved in maximum non-bonding interactions (11) and one hydrogen bond (Table 5). Further, the high absolute values of dock score (-42.70), binding energy (-7.78 kcal/mol), and dissociation constant (pKd, 5.70) also suggested that EGCG bound tightly to NF-κB and thus may act as a potential inhibitor for DNA binding activity.
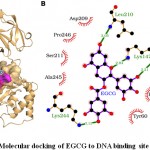 |
Figure 5: Molecular docking of EGCG to DNA binding site of NF-κB. |
Table 5: The transcription factor NF-κB residues interacting with EGCG are listed with the number of non-bonding contacts and hydrogen bonds.
| Interacting residues | Hydrogen bonds | Non-bonded contacts |
| Tyr-60 | 8 | |
| His-144 | 1 | |
| Thr-146 | 3 | |
| Lys-147 | 2 | 8 |
| Asp-209 | 5 | |
| Leu-210 | 2 | |
| Ser-211 | 4 | |
| Lys-244 | 1 | 11 |
| Ala-245 | 2 | |
| Pro-246 | 3 |
Comparative Binding Analyses of Catechin Derivatives
It is noteworthy that all the interacting residues of EC and EGC were common except for one residue of EC, Lys-147 (Table 2) and the binding scores were comparable i.e., dock score (EC, -32.01; EGC, -33.12), binding energy (EC, -7.14 Kcal; EGC, -7.08 Kcal) and pKd (EC, 5.23; EGC, 5.19) indicating towards similarity in binding pose and strength (Table 6). This finding is in agreement with what is expected as the structure of EC and EGC are similar (Fig. 1). Similarly, the binding pattern of ECG and EGCG were found similar as both have same set of ten interacting residues (Table 6) and have comparable binding scores i.e., dock score (ECG, -43.72; EGCG, -42.70), binding energy (ECG, -7.84 Kcal; EGCG, -7.78 Kcal) and pKd (ECG, 5.75; EGCG, 5.70) as shown in Table 2. This property of similarity in binding pattern is also attributed to similarity in structures of ECG and EGCG (Fig. 1). However, the binding affinity order is ‘ECG ≈ EGCG > EGC ≈ EC’ based on the binding scores. Collectively, all the four catechin derivatives have six common interacting residues (Tyr-60, Asp-209, Leu-210, Ser-211, Lys-244 and Pro-246) and interestingly the residue Lys-244 was identified as the key residues in all cases as it was involved in the maximum number of non-bonding interactions. These findings indicated towards overall similarity in the binding of catechin derivatives in the DNA binding site of NF-κB.
Table 6: Comparative binding analyses of catechin derivatives in the binding site of NF-κB. Each column contains the interacting residues with the catechin derivative mentioned on the top of the column. The common interacting residues for different derivatives are placed in the same row.
| EC | EGC | ECG | EGCG |
| Tyr-60 | Tyr-60 | Tyr-60 | Tyr-60 |
| – | – | His-144 | His-144 |
| – | – | Thr-146 | Thr-146 |
| Lys-147 | – | Lys-147 | Lys-147 |
| Asp-209 | Asp-209 | Asp-209 | Asp-209 |
| Leu-210* | Leu-210* | Leu-210* | Leu-210* |
| Ser-211 | Ser-211 | Ser-211 | Ser-211 |
| Lys-244 | Lys-244 | Lys-244 | Lys-244 |
| – | – | Ala-245 | Ala-245 |
| Pro-246 | Pro-246 | Pro-246 | Pro-246 |
| Asn-247 | Asn-247 | – | – |
Conclusion
The molecular docking results suggest that the green tea catechin derivatives, EC, ECG, EGC, and EGCG showed substantial binding to NF-κB and may inhibit DNA binding activity of NF-κB, which may result in inhibition of NF-κB mediated transcription of target genes. The anti-cancer properties of these compounds are well established, and NF-κB is a known therapeutic target. Therefore, this study proposes the catechin derivatives as a potential inhibitor of NF-κB and hence downstream pathways, which may prevent or minimize the onset and progression of various cancer types. The order of their binding affinity in decreasing order based on the dock scores, binding energy and dissociation constant values are as follows: ECG ≈ EGCG > EGC ≈ EC. In general, all these four catechin derivatives have high binding strength and may act as a potential inhibitor of NF-κB mediated carcinogenesis. However, the ECG (the top scorer) and EGCG (2nd rank), having a similar structure, specifically showed high comparative values for the binding affinities. Thus these may be proposed as potentially effective inhibitors of NF-κB for DNA binding activity.
Acknowledgements
The authors are thankful to School of Life & Allied Health Sciences, The Glocal University, Saharanpur, Uttar Pradesh, India for the facilities provided.
Conflict of Interest
The authors confirm that they do not have any conflict of interest.
References
- Global cancer facts & figure. American Cancer Society, 1–60 (2008).
- Baud V and Karin M. Is nf-kappab a good target for cancer therapy? Hopes and pitfalls. Nature Reviews Drug Discovery, 8(1), 33-40 (2009).
- Plewka D, Plewka A, Miskiewicz A, Morek M and Bogunia E. Nuclear factor-kappa b as potential therapeutic target in human colon cancer. Journal of Cancer Research and Therapeutics, 14(3), 516-520 (2018).
- Ghosh S and Karin M. Missing pieces in the nf-kappab puzzle. Cell, 109 Suppl, S81-96 (2002).
- Karin M and Ben-Neriah Y. Phosphorylation meets ubiquitination: The control of nf-[kappa]b activity. Annual Review of Immunology, 18, 621-663 (2000).
- Karin M and Greten FR. Nf-kappab: Linking inflammation and immunity to cancer development and progression. Nature Reviews Immunology, 5(10), 749-759 (2005).
- Kumar A, Takada Y, Boriek AM and Aggarwal BB. Nuclear factor-kappab: Its role in health and disease. Journal of Molecular Medicine (Berlin, Germany), 82(7), 434-448 (2004).
- Prasad S, Ravindran J and Aggarwal BB. Nf-kappab and cancer: How intimate is this relationship. Molecular and Cellular Biochemistry, 336(1-2), 25-37 (2010).
- Sethi G, Ahn KS, Sandur SK, Lin X, Chaturvedi MM and Aggarwal BB. Indirubin enhances tumor necrosis factor-induced apoptosis through modulation of nuclear factor-kappa b signaling pathway. The Journal of Biological Chemistry, 281(33), 23425-23435 (2006).
- Dutta J, Fan Y, Gupta N, Fan G and Gélinas C. Current insights into the regulation of programmed cell death by nf-kappab. Oncogene, 25(51), 6800-6816 (2006).
- Luo J-L, Kamata H and Karin M. Ikk/nf-kappab signaling: Balancing life and death–a new approach to cancer therapy. The Journal of Clinical Investigation, 115(10), 2625-2632 (2005).
- Karin M, Cao Y, Greten FR and Li Z-W. Nf-kappab in cancer: From innocent bystander to major culprit. Nature Reviews Cancer, 2(4), 301-310 (2002).
- Bharti AC and Aggarwal BB. Nuclear factor-kappa b and cancer: Its role in prevention and therapy. Biochemical Pharmacology, 64(5-6), 883-888 (2002).
- Godwin P, Baird AM, Heavey S, Barr MP, O’Byrne KJ and Gately K. Targeting nuclear factor-kappa b to overcome resistance to chemotherapy. Frontiers in Oncology, 3(120 (2013).
- Miller SC, Huang R, Sakamuru S, Shukla SJ, Attene-Ramos MS, Shinn P, Van Leer D, Leister W, Austin CP and Xia M. Identification of known drugs that act as inhibitors of nf-kappab signaling and their mechanism of action. Biochemical Pharmacology, 79(9), 1272-1280 (2010).
- Bhattacharya S, Muhammad N, Steele R, Kornbluth J and Ray RB. Bitter melon enhances natural killer mediated toxicity against head and neck cancer cells. Cancer prevention research (Philadelphia, Pa), 10(6), 337-344 (2017).
- Bhattacharya S, Muhammad N, Steele R, Peng G and Ray RB. Immunomodulatory role of bitter melon extract in inhibition of head and neck squamous cell carcinoma growth. Oncotarget, 7(22), 33202-33209 (2016).
- Muhammad N, Steele R, Isbell TS, Philips N and Ray RB. Bitter melon extract inhibits breast cancer growth in preclinical model by inducing autophagic cell death. Oncotarget, 8(39), 66226-66236 (2017).
- Lichota A and Gwozdzinski K. Anticancer activity of natural compounds from plant and marine environment. International Journal of Molecular Sciences, 19(11) (2018).
- Zaveri NT. Green tea and its polyphenolic catechins: Medicinal uses in cancer and noncancer applications. Life Sciences, 78(18), 2073-2080 (2006).
- Fassina G, Venè R, Morini M, Minghelli S, Benelli R, Noonan DM and Albini A. Mechanisms of inhibition of tumor angiogenesis and vascular tumor growth by epigallocatechin-3-gallate. Clinical Cancer Research, 10(14), 4865-4873 (2004).
- Zhao Y, Yu L, Xu S, Qiu F, Fan Y and Fu G. Down-regulation of connexin43 gap junction by serum deprivation in human endothelial cells was improved by (-)-epigallocatechin gallate via erk map kinase pathway. Biochemical and Biophysical Research Communications, 404(1), 217-222 (2011).
- Reygaert WC. Green tea catechins: Their use in treating and preventing infectious diseases. BioMed Research International, 9105261 (2018).
- Li F, Wang Y, Li D, Chen Y, Qiao X, Fardous R, Lewandowski A, Liu J, Chan T-H and Dou QP. Perspectives on the recent developments with green tea polyphenols in drug discovery. Expert opinion on drug discovery, 13(7), 643-660 (2018).
- Adachi S, Nagao T, Ingolfsson HI, Maxfield FR, Andersen OS, Kopelovich L and Weinstein IB. The inhibitory effect of (-)-epigallocatechin gallate on activation of the epidermal growth factor receptor is associated with altered lipid order in ht29 colon cancer cells. Cancer Research, 67(13), 6493-6501 (2007).
- Naponelli V, Ramazzina I, Lenzi C, Bettuzzi S and Rizzi F. Green tea catechins for prostate cancer prevention: Present achievements and future challenges. Antioxidants (Basel, Switzerland), 6(2) (2017).
- Qiao Y, Cao J, Xie L and Shi X. Cell growth inhibition and gene expression regulation by (-)-epigallocatechin-3-gallate in human cervical cancer cells. Archives of Pharmacal Research, 32(9), 1309-1315 (2009).
- Jamal MS, Parveen S, Beg MA, Suhail M, Chaudhary AGA, Damanhouri GA, Abuzenadah AM and Rehan M. Anticancer compound plumbagin and its molecular targets: A structural insight into the inhibitory mechanisms using computational approaches. PloS One, 9(2), e87309 (2014).
- Mohd Rehan. A structural insight into the inhibitory mechanism of an orally active pi3k/mtor dual inhibitor, PKI-179 using computational approaches. Journal of Molecular Graphics & Modelling, 62, 226-234 (2015).
- Rehan M. An anti-cancer drug candidate OSI-027 and its analog as inhibitors of mtor: Computational insights into the inhibitory mechanisms. Journal of Cellular Biochemistry, 118(12), 4558-4567 (2017).
- Rehan M, Ahmad E, Sheikh IA, Abuzenadah AM, Damanhouri GA, Bajouh OS, AlBasri SF, Assiri MM and Beg MA. Androgen and progesterone receptors are targets for bisphenol a (bpa), 4-methyl-2,4-bis-(p-hydroxyphenyl)pent-1-ene–a potent metabolite of bpa, and 4-tert-octylphenol: A computational insight. PloS One, 10(9), e0138438 (2015).
- Rehan M and Bajouh OS. Virtual screening of naphthoquinone analogs for potent inhibitors against the cancer-signaling pi3k/akt/mtor pathway. Journal of Cellular Biochemistry, (2018).
- Rehan M. Anticancer compound XL765 as PI3K/mTOR dual inhibitor: A structural insight into the inhibitory mechanism using computational approaches. PloS One, 14(6), e0219180 (2019).
- Rehan M and Mostafa M. Virtual screening of 1,4-naphthoquinone derivatives for inhibition of a key cancer signaling protein, akt1 kinase. Anticancer Research, 39(7), 3823-3833 (2019).
- Ewing TJ, Makino S, Skillman AG and Kuntz ID. Dock 4.0: Search strategies for automated molecular docking of flexible molecule databases. Journal of Computer-Aided Molecular Design, 15(5), 411-428 (2001).
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC and Ferrin TE. Ucsf chimera–a visualization system for exploratory research and analysis. Journal of Computational Chemistry, 25(13), 1605-1612 (2004).
- DeLano WL. The pymol molecular graphics system. Proteins, 30, 442-454 (2002).
- Laskowski RA and Swindells MB. Ligplot+: Multiple ligand-protein interaction diagrams for drug discovery. Journal of Chemical Information and Modeling, 51(10), 2778-2786 (2011).
- Wang R, Lai L and Wang S. Further development and validation of empirical scoring functions for structure-based binding affinity prediction. Journal of Computer-Aided Molecular Design, 16(1), 11-26 (2002).

This work is licensed under a Creative Commons Attribution 4.0 International License.





