Manuscript accepted on : 22-May-2019
Published online on: 27-05-2019
Plagiarism Check: Yes
Reviewed by: Ebrahim Kouhsari
Second Review by: Francisco Solano
Final Approval by: Prof. Giacomo Cao
Molecular Study of E. coli Virulence Genes in Nosocomial Sepsis
Maysaa E. Zaki*1 , Samah Bastawy2 and Karim Montasser2
, Samah Bastawy2 and Karim Montasser2
1Clinical Pathology, Mansoura Faculty of Medicine, Egypt.
2Clinical Pathology, Helwan Faculty of Medicine, Egypt.
Corresponding Author E-mail: may_s65@hotmail.com
DOI : http://dx.doi.org/10.13005/bbra/2743
ABSTRACT: Escherichia coli (E. coli) is a common cause of nosocomial sepsis. There are multiple factors related to the severity of sepsis among these are the presence of virulence genes and the pattern of antibiotics resistance. The aim of the present study was to determine the prevalence of virulence pap gene encoding for pili, hlyA gene encoding for α-hemolysin and cnf1 gene encoding for cytotoxic necrotizing factor 1 among E. coli isolated from children with nosocomial sepsis. Also, to correlate the presence of ESBL and carbapenem resistance with the presence of these genes. The study is a retrospective cross-sectional study included 150 non-duplicate strains of E. coli isolated from blood cultures from children with nosocomial sepsis. The isolated E. coli strains were subjected to antibiotics study by disc diffusion method, detection of extended spectrum lactamase production by double discs diffusion method and determination of resistance to carbapenem by combined tests methods. The detection of virulence genes pap, hylA and cnf-1 were determined by multiplex polymerase chain reaction (PCR). E. coli isolates were classified as ESBL phenotype in 56% of the isolates and carbapenemase producing phenotype in 34.7%. Pap gene, hylA and cnf-1 genes were detected in 30%, 23.3% and 22.7% of the isolated E. coli. The clinic-laboratory study of the virulence genes of E. coli revealed the significant association of pap, hylA and cnf-1genes with prolonged duration of the use of the medical devices (4.3± 2.9 days-P=0.01, 4.5± 2.9 days, P=0.02, 5.2± 3.4 days, P=0.0001 respectively). HylA gene was associated with younger age of the patients (28.4± 4.5, P=0.01). Pap gene was significantly associated with ESBLs and carbapenemase phenotypes (P=0.0001, P=0.002 respectively). On the other hand, cnf-1 was significantly associated with E. coli isolated from primary sepsis (P=0.02) and in isolates from sepsis due to medical devices (P=0.02) and was significantly associated with death (P=0.01) and carbapenemase resistance (P=0.01). The present study highlights the prevalence of pap, hylA and cnf-1 virulence genes among E. coli associated with nosocomial sepsis in children. The frequency of some of these genes was correlated with extended spectrum lactamase resistance and carbapenemase resistance. This may be attributed to the presence of the virulence and antibiotics genes on transferable plasmids. Moreover, there was association with cnf-1 virulence gene and mortality outcome of sepsis. Further studies are recommended to evaluate these findings.
KEYWORDS: Cnf-1; E. Coli; HylA; Multiplex PCR; Pap
Download this article as:| Copy the following to cite this article: Zaki M. E, Bastawy S, Montasser K. Molecular Study of E. coli Virulence Genes in Nosocomial Sepsis. Biosci Biotech Res Asia 2019;16(2). |
| Copy the following to cite this URL: Zaki M. E, Bastawy S, Montasser K. Molecular Study of E. coli Virulence Genes in Nosocomial Sepsis. Biosci Biotech Res Asia 2019;16(2). Available from: https://bit.ly/2Qtqt8R |
Introduction
Sepsis is a serious infection affecting children admitted to intensive care units. There are various pathogens associated with this infection either gram negative bacilli, gram positive cocci and fungal pathogens. Escherichia coli (E. coli) represents a major etiology of sepsis (1,2). Sepsis due to E. coli can result as a complication of infections of urinary tract or gastrointestinal tract. It may also arise as a primary blood stream infection without an obvious source (3).
Various factors are correlated with the severity of E. coli associated with sepsis. Among these factors are resistance pattern to antibiotics that has emerged among E. coli. The isolated E. coli from sepsis has been shown to produce Extended spectrum β-lactamases (ESBLs) (4, 5). Another threat of antibiotics resistance among clinical isolates of E. coli is the resistance to carbapenem antibiotics namely to meropenem and/or imipenem (6). The risks factors for acquiring nosocomial sepsis due to ESBLs strains of E. coli and carbapenem resistant strains include the duration of intensive care units stay, age of the patients, presence of co morbidities and associated devices (4,7).
Other factor that determines the severity of sepsis due to E. coli is the presence of the virulence genes. Virulence genes of E. coli act through several mechanisms. First mechanism is associated with invasion of blood stream controlled by genes that encode for amyloid curli and gene of siderophores that support survival outside gastrointestinal tract (8). Second mechanism is by facilitating the attachment of E. coli to cells by the means of fimbria factors P and S and causing damage to these cells by hemolysin and cytotoxin necrotizing factors (9, 10). Third mechanism is associated with the presence of capsular K1 that inhibits phagocytosis and complement mediated-killing (11,12). The fourth mechanism is by the production of bacteriocins by certain E. coli strains (13).
The aim of the present study was to determine the prevalence of virulence pap gene encoding for pili, hlyA gene encoding for α-hemolysin and cnf1 gene encoding for cytotoxic necrotizing factor 1 among E. coli isolated from children with nosocomial sepsis. Also, to correlate the presence of ESBLs and carbapenem resistance with the presence of these genes.
Materials and Method
The study is retrospective cross-sectional study included 150 non-duplicate strains of E. coli isolated from blood cultures from children with nosocomial sepsis admitted to ICUs in Mansoura University Children hospital, Egypt from January 2016 till January 2018. The study was approved by Mansoura Faculty of Medicine ethical committee and approval consent was obtained from the parents of each child.
The clinical data of each participating child was obtained as regard age, sex, the presence of comorbidities conditions, underlying medical conditions, the presence of central venous line, urinary catheter and other devices and the outcome of infection after 30 days was recorded either death or discharge. The bacteremia was defined as secondary if the cause of sepsis was identified according to clinical and/or laboratory evidence of infection and primary if there was no evidence of the infection
Positive blood culture from defined nosocomial sepsis which is defined as the infection that occurred after 48hours from hospital admission according to CDC definitions was processed according to the standard microbiological methods. Blood culture was obtained under complete sterile conditions and inoculated to blood culture bottles Bact/alert system. Positive blood culture was processed by subculture on blood agar plates and incubated at 37°C for 24 hours. The culture was identified by gram stain followed by biochemical identification by the use of automated microbiology system Microscan (WalkAway 40 plus System- B1018-283- Beckman Coulter, Inc). Antimicrobial susceptibility testing was performed by the use of the Kirby–Bauer disc diffusion method. Production of ESBLs was confirmed using the double-disk synergy test in accordance with the Clinical and Laboratory Standards Institute standards (14). Carbapenem producing E. coli strains were defined as resistant strains to imipenem and/or meropenem discs and confirmed by combined discs diffusion method by the use of EDTA and boronic acid.
Antibiotics Discs diffusion
The used antibiotics discs were ceftazidime (30 μg), cefotaxime (30 μg), cefepime (30 μg), imipenem (10 μg), meropenem (10 μg), gentamicin (10 μg), amikacin (30 μg), ciprofloxacin (5 μg), levofloxacin (5 μg), sulfamethoxazole/trimethoprim(1.25/23.75 μg), and piperacillin/tazobactam (100 μg/10 μg) (Oxoid-Thermo Fisher Scientific-USA 02451). The interpretation of the results was done according to the Clinical and Laboratory Standards Institute (CLSI) guidelines (14).
Double Discs Diffusion Method (DDT) for ESBLs
Resistant strains of E. coli for ceftazidime and cefotaxime antibiotics discs were used for confirmatory phenotypic tests for EDBLs production by DDT. The test depends upon increase the inhibition diameter zone around each disc when antibiotics were conjugated with clavulanic acid a known inhibitory for extended spectrum β-lactamase enzyme by ≥5 mm. The test was performed as previously described (14).
Determination of Carbapenemases Production by Combined Disk Test (CDT) and Boronic Acid Discs
E.coli strains with resistance to imipenem and/or to meropenem were subjected to determination of carbapenemases by CDT and boronic acid discs amino phenylboronic acid (APBA). The positive CDT detects the possible production of metallo-β-lactamase (MBL) and the positive boronic acid disc detects the possible presence of class A carbapenemase (14). The increase of the inhibitory zone around carbapenem discs by ≥5 mm when combined β-lactamase inhibitor (APBA or EDTA) was considered to be a positive result (16).
Quality Control
E. coli ATCC 25922 was used as a susceptible strain for antibiotics susceptibility tests. ESBL-positive Klebsiella pneumoniae ATCC 700603 and ESBL-negative E. coli ATCC 25922 control strains were used in this study. For carbapenemase detection, E. coli ATCC 25922 strain was used as a negative control.
Molecular Study of E. coli
Extraction of E. coli DNA
E. coli was subculture on nutrient agar at 37°C for 24 hours. The colonies were obtained and suspended in Tris-EDTA buffer (10 mMTris-HCI and 1 mM EDTA) and subjected to centrifugation at 2000 g for 10 min. The supernatant was removed and the pellet was incubated with proteinase K and Tris-EDTA buffer for 18 hours at 55°C in water bath. Then, the DNA was extracted by phenol and chloroform extraction method and suspended in Tris-EDTA buffer and kept frozen at -20°C for polymerase chain reaction (PCR) (15).
PCR for Identification of E. coli
The isolates were confirmed as E. coli by detection of gene of Enterobacteriaceae common antigen (ECA) specific for E. coli. PCR method. The used specific primers for E. coli were listed in table 1. (15). The amplification was performed by the use of Qiagen amplification mixture with total volume 50lµ (Qiagen-Germany). The amplification procedures were initial denaturation at 95°C for 7 min; followed by 35 cycles of denaturation at 95°C for 1.5 min, annealing at 65°C for1.5 min and extension at 72°C for 1.5 min and a final extension for 7 min at 72°C. Ten microliters of the reaction mixture was analyzed on 1.5% agarose gel by electrophoresis. The bands were visualized by ethidium bromide staining under UV.
Multiplex PCR for Detection of Virulence Genes
The multiplex PCR was performed by the use of ready to use Qiagen amplification mixture with total volume 50 µl. The sequences of the used primers were summarized in table 1. The amplification procedures were initial denaturation at 94°C for 3 min, followed by 30 cycles of denaturation at 94°C for 1 min, annealing at 63°C for 30 s and extension at 72°C for 3 minutes and the final extension for 7 minutes at 72°C. A volume of 10 ml of the reaction mixture was analyzed on 2% agarose gel by electrophoresis. Bands were visualized under UV after ethidium bromide staining (15).
Table 1: Genes, primers sequences and amplification products bp.
| Gene | Sequences of the Primers | bp |
| E.col | 5’-GGT GGTCGG CAA GCT TTA TCT CAGGGT-3’ 5’-TAA ATT GGG GCT GCC ACC ACG-3’ |
793 |
| papG
|
5’-GCA ACA GCA ACG CTG GTT GCA TCA T-3’
5’-AGA GAG AGC CAC TCT TAT ACG GAC A-3’ |
336 bp |
| hlyA 5’ | 5’-AAC AAG GAT AAG CAC TGT TCT GGC T-3’ 5’-ACC ATA TAA GCG GTC ATT CCC GTC A-3’ |
1177 bp |
| cnf1 | 5’-AAG ATG GAG TTT CCT ATG CAG GAG-3’ 5’-CAT TCA GAG TCC TGC CCT CAT TAT T-3’ |
498 bp |
Statistical Analysis
Data were analyzed by the use of statistical package for social science (SPSS) version 24. The quantitative data were presented as mean, standard deviations and ranges. The qualitative data were presented as percentage and compared by the use of chi-square. The comparison between the studied groups were done by using One Way Analysis of Variance (ANOVA). P was considered significant if ≤0.05. Multiple logistic regression was performed by the use of binary model.
Result
The study included non-replicated isolates of E. coli. The isolates were from children with nosocomial sepsis with median age 6.7 months and they were mainly males 53.3%. The sepsis was primary sepsis in 83.3% of the patients associated with devices central venous catheter, urinary catheter and ventilator (64%). The mortality among these series of patients was 31.3%. E. coli isolates had ESBLs phenotype in 56% of the isolates and carbapenemase producing phenotype in 34.7%. Pap gene, hylA and cnf-1 genes were detected in 30%, 23.3% and 22.7% of the isolated E. coli, table 2.
Table 2: Demographic, clinical and laboratory data of the studied patients.
| Gender | |
| Male | 80 53.3% |
| Female | 70 46.7% |
| Age (months) | |
| Minimum | 1 |
| Maximum | 192 |
| Median | 6.7 |
| Type of Sepsis | |
| Primary | 125 83.3% |
| Secondary | 25 16.7% |
| Type of Sepsis | |
| Device associated | 96 64% |
| Non-device associated | 54 36% |
| Central venous catheter | 72 48% |
| Urinary catheter | 44 29.3% |
| Ventilator | 24 16% |
| Virulence genes | |
| Pap | 45 30% |
| hlyA | 35 23.3% |
| cnf-1 | 34 22.7% |
| ESBLs | 84 56% |
| Carbapenemase | 52 34.7% |
| Outcome | |
| Death | 47 31.3% |
| Survive | 103 68.7% |
The clinic-laboratory study of the virulence genes of E. coli revealed the significant association of pap, hylA and cnf-1 genes with prolonged duration of the use of the medical devices (4.3± 2.9 days-P=0.01, 4.5± 2.9 days, P=0.02, 5.2± 3.4 days, P=0.0001 respectively). HylA gene was associated with younger age of the patients (28.4±4.5, P=0.01). Pap gene was significantly associated with ESBLs and carbapenemase phenotypes (P=0.0001, P=0.002 respectively). On the other hand, cnf-1 was significantly associated with E. coli isolated from primary sepsis (P=0.02) and in isolates from sepsis due to medical devices (P=0.02) and was significantly associated with death (P=0.01) and carbapenemase resistance (P=0.01), table 3.
Table 3: Clinco-laboratory study of the presence of virulence genes of E.
| Pap | hlyA | cnf-1 | |
| (n=45) | (n=35) | (n=34) | |
| Geneder | |||
| Male | 22 48.9% |
20 57.1% |
16 47.1% |
| Female | 23 51.1% |
15 42.9% |
18 52.9% |
| P | 0.3 | 0.4 | 0.3 |
| Age | |||
| Present | 55.8±37.3 | 28.4±4.5 | 52.2±33.3 |
| Absent | 52.7±32.9 | 72.3±5.4 | 57.5±33.4 |
| P | 0.1 | 0.01 | 0.9 |
| Type of sepsis | |||
| Primary | 35 77.8% | 32 91.4% | 24 70.6% |
| Secondary | 10 22.2% | 3 8.6% | 10 29.4% |
| P | 0.2 | 0.1 | 0.02 |
| CVC | 22 48.9% |
19 54.3% |
16 47.1% |
| P | 0.5 | 0.4 | 0.5 |
| Urinary Catheter | 14 31.1% |
9 25.7% |
10 29.4% |
| P | 0.8 | 0.4 | 0.5 |
| Device associated | 20 44.4% |
17 48.6% |
21 61.8% |
| 0.1 | 0.1 | 0.04 | |
| Duration of the presence of the device till sepsis (mean± SD) days | 4.3± 2.9 | 4.5± 2.9 | 5.3± 3.4 |
| Absent | 3.1±2.6 | 2.5±1.1 | 2.9±2.2 |
| P | 0.01 | 0.02 | 0.0001 |
| ESBLs | 42 93.3% |
26 74.3% |
22 64.7% |
| P | 0.0001 | 0.02 | 0.2 |
| Carbapenemase | 24 53.3% |
12 34.3% |
18 52.9% |
| P | 0.002 | 0.5 | 0.01 |
| Outcome | |||
| Death | 14 31.1% |
13 37.4% |
30 88.2% |
| Life | 31 68.9% |
22 62.9% |
4 11.8% |
| P | 0.6 | 0.3 | 0.01 |
Multiple logistic regression analysis of possible risk factors associated with death reveals significant association with younger age and the presence of cnf-1 as significant risk factors (P=0.012, P=0.004 respectively), table 4.
Table 4: Multiple logistic regression study of risk factors associated with death.
| B | Wald | Sig. | Exp(B) | 95% C.I. | ||
| Lower | Upper | |||||
| age | 0.012 | 6.364 | 0.012 | 1.012 | 1.003 | 1.022 |
| gender | 0.514 | 1.671 | 0.196 | 1.671 | 0.767 | 3.641 |
| ESBLS | -0.616 | 1.882 | 0.17 | 0.54 | 0.224 | 1.302 |
| EDT | 0.431 | 1.036 | 0.309 | 1.539 | 0.671 | 3.531 |
| IV_Days | -0.005 | 0.004 | 0.952 | 0.995 | 0.856 | 1.157 |
| Device_associated | 0.088 | 0.058 | 0.81 | 1.092 | 0.532 | 2.242 |
| pap | 0.06 | 0.013 | 0.909 | 1.062 | 0.383 | 2.941 |
| hly | -0.118 | 0.053 | 0.817 | 0.889 | 0.326 | 2.422 |
| cnf1 | -1.436 | 8.172 | 0.004 | 0.238 | 0.089 | 0.637 |
The isolated E. coli had high prevalence of resistance toward ceftazidime, meropenem and imipenem (73.2%, 58.7%, 53.3% respectively) with lower prevalence of resistance to cefepime, amikacin and ciprofloxacin (17.3%, 13.3%, 20% respectively), figure 1.
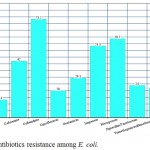 |
Figure 1: Antibiotics resistance among E. coli.
|
Discussion
E.coli has different strains that can act as a commensal in gastrointestinal tract, diarrheagenic bacteria or an extraintestinal tract pathogen. It is a common cause of nosocomial infections with presence of several virulence genes that are not present in the commensal strains (16). Beside virulence factors associated with pathogenic E. coli, antibiotics resistance seems to play a vital role in its association with severe infections such nosocomial sepsis.
 |
Figure 2: Positive E. coli for pap genes M: marker Lanes 1-4 positive E. coli for pap gene.
|
In the present report the prevalence of the virulence genes pap, cnf-1 and hylA were studied in correlation with antibiotics resistance and outcome of nosocomial sepsis in children.
The frequency of cnf-1 gene among clinical isolates of E. coli was 22.7%. This frequency is online with findings of previous studies with range from 14.2% up to 30% (17-19). The frequency was less in commensal isolates. Moreover, cnf-1 gene was the only virulent gene reported in the present study to be correlated with mortality (P=0.004). There are scarce data about association of cnf-1 gene with outcome of sepsis in previous studies (18, 19) with a suggestion that there is a link between fatal outcome and the presence of this gene. This link may be attributed to the fact that cnf-1 codes for cytotoxic necrotizing factor 1 that affects the host cellular immune functions leading to inhibition of phagocytosis and chemotactic activities of neutrophils (20, 21).
The mortality rates in the present series of patients was 31.1% which is an expected rate as the sepsis was mainly primary sepsis in 83.3% of the patients associated with the use of medical devices in 64%. The mortality rates in previous reports range from 5% up to 30% in sepsis due to non-urinary E. coli sepsis (19, 22).
The frequency of pap gene was 30% with non-significant correlation with mortality outcome. Previous data supported the present finding with pap gene the most frequent gene associated with E. coli as it encodes for the P fimbriae that mediates an important function of bacterial adherence to host cells (23). However, its relation to outcome of sepsis had significant variance in the studies, as one study reported an association to fatal outcome (18) while other study claimed a protective role for this gene from fatal outcome due to the immunogenicity of the P fimbriae leading to early and strong immune response that controls the sepsis (24).
The frequency of hyla gene was 23.3%. Hemolysin (hly) is produced by various pathogenic types of E. coli causing extraintestinal and intestinal infections, but its effect on virulence is not completely clarified (25,26). HlyA gen region was related with complicated urinary tract infections such as pyelonephritis and cystitis (27, 28).
The frequency of detections of pap, hylA and cnf-1 genes were correlated with the prolonged duration of the use of the medical devices (P=0.01, P=0.02, P=0.0001 respectively). Previous reports confirmed the correlation between device associated infections and virulence genes due to increase capacity of E. coli to form biofilm difficult to treat (16, 29). From the present study this capacity may be increased by prolonged use of medical devices such as central venous catheter, ventilator and urinary catheter.
The pap gene was correlated with ESBLs and carbapenemase resistant phenotypes and cnf-1 was correlated with carbapenemase phenotype. The correlation between virulence genes and antibiotics resistance in E. coli are controversy, while some studies reported this association with ESBLs (30, 31) other study reported no correlation (32). There are many theories about the association between antibiotics resistance and virulence genes one theory attributed this association due to the presence of both virulence genes and antibiotics resistance genes on the same plasmid while the other theory related this to porin loss (33), and modifications in the penicillin binding proteins and efflux pumps mechanism (34). Efflux pumps are responsible for the secretion of the molecules containing virulence factors that is regulated by quorum sensing which has a positive effect on antibiotics resistance and virulence (35,36).
The isolated E. coli had high prevalence of resistance toward ceftazidime, meropenem and imipenem with lower prevalence of resistance to cefepime, amikacin and ciprofloxacin.The high frequency of antibiotics resistance among E.coli may be attributed to the improper use of antibiotics with selective pressure leading to the prevalence of resistant strains (37). This finding warrants the need for strict implantation of antibiotics policy.
The present study highlights the prevalence of pap, hylA and cnf-1 virulence genes among E. coli associated with nosocomial sepsis in children. The frequency of some of these genes was correlated with extended β- spectrum lactamase resistance and carbapenemase resistance. This may be attributed to the presence of the virulence and antibiotics genes on transferable plasmids. Moreover, there was association with cnf-1 virulence gene and mortality outcome of sepsis. Further studies are recommended to evaluate these findings.
Acknowledgments
The authors would like to thank all support staff and participating patients in this study.
Conflicts of Interest
There is no conflicts of interest.
Funding source
There is no funding source.
References
- Geerdes HF, Ziegler D, Lode H, Hund M, Loehr A, Fangmann W, et al. Septicemia in 980 patients at a university hospital in Berlin: Prospective studies during 4 selected years between 1979 and 1989. Clin Infect Dis 1992; 15:991-1002.
CrossRef - Reddy, E.A., Shaw, A.V., Crump, J.A., 2010. Community-acquired bloodstream infections in Africa: a systematic review and meta-analysis. Lancet Infect. Dis. 10(6), 417-432.
CrossRef - Laupland, K.B., Gregson, D.B., Church, D.L., Ross, T., Pitout, J.D., 2008.Incidence, risk factors and outcomes of Escherichia coli bloodstream infections in a large Canadian region.Clin.Microbiol. Infect. 14(11), 1041-1047.
CrossRef - Denis B, Lafaurie M, Donay JL, et al. Prevalence, risk factors, and impact on clinical outcome of extended-spectrum beta-lactamase-producing Escherichia coli bacteraemia: a five-year study. Int J Infect Dis 2015; 39:1–6.
CrossRef - Cornejo-Juárez P, Pérez-Jiménez C, Silva-Sánchez J, et al. Molecular analysis and risk factors for Escherichia coli producing extended-spectrum β-lactamase bloodstream infection in hematological malignancies. PLoS One 2012;7:e35780
McLaughlin M, Advincula MR, Malczynski M, Qi C, Bolon M, Scheetz MC. Correlations of Antibiotic Use and Carbapenem Resistance in Enterobacteriaceae, Antimicrobial Agents and Chemotherapy 2013;57:5131-3.
CrossRef - Rodríguez-Baño J, Navarro MD, Romero L, et al. Risk-factors for emerging bloodstream infections caused by extended-spectrum beta-lactamase-producing Escherichia coli. Clin Microbiol Infect 2008;14(2):180–3.
CrossRef - Hung, C., Marschall, J., Burnham, C.A., Byun, A.S., Henderson, J.P., 2015. The bacterial amyloid curli is associated with urinary source bloodstream infection. PLoS One.9(1), e86009.
CrossRef - Johnson, J.R., 1991. Virulence factors in Escherichia coli urinary tract infection. Clin.Microbiol. Rev. 4(1), 80-128.
CrossRef - Mulvey, M.A., 2002.Adhesion and entry of uropathogenic Escherichia coli.Cell Microbiol. 4(5), 257-71.
CrossRef - Gaschignard, J., Levy, C., Romain, O., Cohen, R., Bingen, E., Aujard, Y., Boileau, P., 2011. Neonatal Bacterial Meningitis: 444 Cases in 7 Years. Pediatr. Infect. Dis. J. 30(3), 212-217.
CrossRef - Stoll, B.J., Hansen, N.I., Sanchez, P.J., Faix, R.G., Poindexter, B.B., Van Meurs, K.P., Bizzarro, M.J., Goldberg, R.N., Frantz, I.D.III., Hale, E.C., Shankaran, S., Kennedy, K., Carlo, W.A., Watterberg, K.L., Bell, E.F., Walsh, M.C., Schibler, K., Laptook, A.R., Shane, A.L., Schrag, S.J., Das, A., Higgins, R.D., 2011. Early onset neonatal sepsis: The burden of group B streptococcal and E. coli disease continues. Pediatrics. 127(5), 817–826.
CrossRef - Micenková, L., Bosák, J., Štaudová, B., Kohoutová, D., Čejková, D., Woznicová, V., Vrba, M., Ševčíková, A., Bureš, J., Šmajs, D., 2016.Microcin determinants are associated with B2 phylogroup of human fecal Escherichia coli isolates. Microbiology Open. 5(3), 490-498.
CrossRef - Clinical and Laboratory Standards Institute.Performance Standards for Antimicrobial Susceptibility Testing, 24th Informational Supplement (M100-S24). Wayne, PA: Clinical and Laboratory Standards Institute; 2014:34(1).https://clsi.org/standards/products/microbiology/documents/m100/
- Arisoy M, Aysev D, Ekim M, Ozel D, Köse SK, Ozsoy ED, Akar N. Detection of virulence factors of Escherichia coli from children by multiplex polymerase chain reaction. Int J ClinPract. 2006;60(2):170-3.
CrossRef - Naves P, del Prado G, Huelves L, Gracia M, Ruiz V, Blanco J, Dahbi G, Blanco M, Ponte Mdel C, Soriano F. Correlation between virulence factors and in vitro biofilm formation by Escherichia coli strains. MicrobPathog. 2008 ;45(2):86-91. doi: 10.1016/j.micpath.2008.03.003.
CrossRef - Lefort A, Panhard X, Clermont O, Woerther PL, Branger C, Mentré F, Fantin B, Wolff M, Denamur E; COLIBAFI Group. Host factors and portal of entry outweigh bacterial determinants to predict the severity of Escherichia coli bacteremia. J ClinMicrobiol 2011; 49:777-83; PMID:21177892; http://dx.doi.org/10.1128/JCM.01902-10.
CrossRef - Jauréguy F, Carbonnelle E, Bonacorsi S, Clec’h C, Casassus P, Bingen E, Picard B, Nassif X, Lortholary O. Host and bacterial determinants of initial severity and outcome of Escherichia coli sepsis. ClinMicrobiol Infect 2007; 13:854-62; PMID:17617183.
CrossRef - Mora-Rillo M, Fernández-Romero N, Francisco CN-S, et al. Impact of virulence genes on sepsis severity and survival in Escherichia coli bacteremia. Virulence. 2015;6(1):93-100. doi:10.4161/21505594.2014.991234.
CrossRef - Ananias M, Yano T. Serogroups and virulence genotypes of Escherichia coli isolated from patients with sepsis. Brazilian J Med Biol Res 2008; 41:877-83; PMID:19030710; http://dx.doi.org/10.1590/S0100-879X2008001000008.
CrossRef - Johnson JR, Stell AL. Extended virulence genotypes of Escherichia coli strains from patients with urosepsis in relation to phylogeny and host compromise. J Infect Dis 2000; 181:261-72; PMID:10608775; http://dx.doi.org/10.1086/315217.
CrossRef - Laupland KB, Gregson DB, Church DL, Ross T, Pitout JDD. Incidence, risk factors and outcomes of Escherichia coli bloodstream infections in a large Canadian region. ClinMicrobiol Infect 2008; 14:1041-7; PMID:19040476; http://dx.doi.org/10.1111/j.1469-0691.2008.02089.x
CrossRef - Wiles T, Kulesus R, Mulvey MA. Origins and virulence mechanisms of uropathogenic Escherichia coli. Exp Mol Pathol2008; 85:11-9; PMID:18482721; http://dx.doi.org/10.1016/j.yexmp.2008.03.
CrossRef - Rodríguez-Baño J, Mingorance J, Fernández-Romero N, Serrano L, López-Cerero L, Pascual A; ESBL-REIPI Group. Outcome of bacteraemia due to extended-spectrum β-lactamase-producing Escherichia coli: impact of microbiological determinants. J Infect 2013; 67:27-34; PMID:23588104; http://dx.doi.org/10.1016/j.jinf.2013.04.006.
CrossRef - Mainil J. Escherichia coli virulence factors. Veterinary Immunology and Immunopathology 2013; 152:2-12.
CrossRef - Bien J, Sokolova O, Bozko P. Role of Uropathogenic Escherichia coli Virulence Factors in Development of Urinary Tract Infection and Kidney Damage. International Journal of Nephrology 2012;681473.
CrossRef - Tarchouna M, Ferjani A, Ben-Selma W, Boukadida J. Distribution of uropathogenic virulence genes in Escherichia coli isolated from patients with urinary tract infection. International Journal of Infectious Diseases 2013;17:e450-3.
CrossRef - Wang MC, Tseng CC, Chen CY, Wu JJ, Huang JJ.The role of bacterial virulence and host factors in patients with Escherichia coli bacteremia who have acute cholangitis or upper urinary tract infection. Clinical Infectious Diseases 2002; 15:1161-6.
CrossRef - Soto SM, Smithson A, Martinez JA, Horcajada JP, Mensa J, Vila J. Biofilm formation in uropathogenic Escherichia coli strains: relationship with prostatitis, urovirulence factors and antimicrobial resistance. J Urol 2007; 177:365–8.
CrossRef - Pitout JDD, Laupland KB, Church DL, Menard ML, Johnson JR. Virulence Factors of Escherichia coli Isolates That Produce CTX-M-Type Extended-Spectrum β-Lactamases. Antimicrob. Agents Chemother. 2005; 49:4667-70.
CrossRef - Sharma S, Bhat GK, Shenoy S. Virulence factors and drug resistance in Escherichia coli isolated from extraintestinal infections. Indian Journal of Medical Microbiology 2007; 25:369-73.
CrossRef - Candan E D ,Aksöz N. Escherichia Coli: Characteristics of Carbapenem Resistance and Virulence Factors. Braz. arch. biol. technol. vol.60 Curitiba 2017. http://dx.doi.org/10.1590/1678-4324-2017160416.
CrossRef - Johnson JR, Stell AL. Extended virulence genotypes of Escherichia coli strains from patients with urosepsis in relation to phylogeny and host compromise. J Infect Dis 2000; 181:261–72.
CrossRef - Nicolas-Chanoine MH, Blanco J, Leflon-Guibout V, Demarty R, Alonso MPM, Caniça M, et al. Intercontinental emergence of Escherichia coli clone O25: H4-ST131 producing CTX-M-15. J AntimicrobChemother 2007; 61:273–81.
CrossRef - Yang HH, Vinopal RT, Grasso D, Smets BF. High diversity among environmental Escherichia coli isolates from a bovine feedlot. Appl Environ Microbiol 2004; 70:1528–36.
CrossRef - Harrison JJ, Ceri H, Yerly J, Stremick CA, Hu Y, Martinuzzi R, et al. The use ofmicroscopy and three-dimensional visualization to evaluate the structure of microbial biofilms cultivated in the Calgary Biofilm Device. Biol Proced Online. 2006; 8:194–215.
CrossRef - Olorunmola FO, Kolawole DO, Lamikanra A. Antibiotic Resistance and Virulence Properties in Escherichia Coli Strains from Cases of Urinary Tract Infections. African Journal of Infectious Diseases. 2013;7(1):1-7.
CrossRef

This work is licensed under a Creative Commons Attribution 4.0 International License.





