Manuscript accepted on : February 19, 2009
Published online on: 22-06-2009
First Report of Bovine Rotavirus From Kerala
R. Ambily2*, M. Mini1, K. John2 G. K. Nair2
1Department of Veterinary Microbiology, College of Veterinary and Animal Sciences, Pookot, Lakkidi P.O., Wayanad - 673 576 India.
2Department of Veterinary Microbiology, College of Veterinary and Animal Sciences, Mannuthy, Kerala - 680 651 India.
Corresponding Author E-mail: ambilysathiadevan@yahoo.co.in
ABSTRACT: A study was conducted to detect the prevalence of rotavirus in diarrhoeic calves in Thrissur district, Kerala. Some of the cattle herds had a history of recurrent diarrhoea. The methods adopted were Reverse Transcriptase-Polymerase Chain Reaction (RT-PCR), Ribonucleic Acid –Poly Acrylamide Gel Electrophoresis (RNA-PAGE) followed by silver staining and Agar Gel Immuno Diffusion (AGID). Among 124 diarrheic faecal samples collected during a period of two years, 29 (23.39 per cent) were detected as positive by RNA-PAGE, 35 (28.23 per cent) samples by RT-PCR and 16 (12.90 per cent) by AGID. The clustered arrangement of the 11 segments of the genome showed a 4:2:3:2 migration pattern, typical of group A bovine rotavirus. RT-PCR was conducted using genome segment 4 (VP4 gene) specific primers. Among the various tests employed, RT-PCR was found to be more sensitive in the diagnosis of Bovine rota viral (BRV) infections. It was found that the occurrence of infection was most common in zero to two weeks of age (39.39 per cent) followed by two to four weeks of age (37.04 per cent).
KEYWORDS: Bovine rota virus; calves; silver staining; RT-PCR
Download this article as:| Copy the following to cite this article: Ambily R, Mini M, John K, Nair G. K. First Report of Bovine Rotavirus From Kerala. Biosci Biotechnol Res Asia 2008;6(1) |
| Copy the following to cite this URL: Ambily R, Mini M, John K, Nair G. K. First Report of Bovine Rotavirus From Kerala. Biosci Biotechnol Res Asia 2008;6(1). Available from: https://www.biotech-asia.org/?p=8139 |
Introduction
The role of rotavirus as a cause of diarrhoea in calves was first demonstrated by Mebus et al.1 at Nebraska Agricultural Experiment Station. The name given was Nebraska Calf Scours Virus, later called as Rotavirus. It is now recognized as the single most important viral agent responsible for gastroenteritis in calves. Several works were carried out in this aspect in different parts of India. However, incidence of bovine rotavirus (BRV) in Kerala has not been studied. Hence, this study was undertaken to detect rota virus in faecal samples of diarrhoeic calves in Kerala. The methods adopted were Reverse Transcriptase-Polymerase Chain Reaction (RT-PCR), RNA- PolyAcrylamide Gel Electrophoresis (RNA-PAGE) and Agar Gel Immunodiffusion (AGID). The influence of age on the occurrence of the infection was also studied.
Materials and Methods
Collection of samples
Faecal samples were collected from 124 diarrhoeic calves of different age groups from Veterinary Hospitals and farms attached to Kerala Agricultural University, various organized dairy farms and animals reared by some individual farmers in Thrissur district, Kerala. Collection of samples was made using dry sterile cotton swabs and was immersed in sterile phosphate buffered saline (PBS, pH 7.2). Freshly voided faeces were also collected and transferred immediately to the laboratory in an ice box and preserved at -20°C until use.
Extraction of RNA
The RNA was extracted from the diarrhoeic faecal samples using TRIZOL reagent as per the manufacturer’s protocol, Molecular Research Centre Inc., Cincinnati (1995)2. All the micro tips and eppendorf tubes used were washed with Di ethyl pyrocarbonate (DEPC) treated water to make it RNase free.
Bovine group A rotavirus RuBV128 kindly provided by Dr. T. N. Naik, Deputy Director and Head, National Centre for Cholera and Enteric Diseases, Kolkata was used as positive sample in the whole study.
Electrophoresis of Rota viral RNA
The segmented RNA genome was analyzed by RNA-PAGE using the discontinuous buffer system as described by Herring et al. 3 followed by silver staining. The extracted RNA was loaded into eight per cent polyacrylamide gels of one millimeter thickness and subjected to electrophoresis at 100 V and 20 mA for 16 h. The gel was stained using silver nitrate solution (0.011M).
Reverse Transcriptase –Polymerase Chain Reaction
In RT-PCR, consensus primers for an 877 bp nucleotide sequence of the VP4 gene of bovine rotavirus were used.
Primer 1 (Forward) Con 3 (11-32) 5”-TGGCTTCGCCATTTTATAGACA-3’
Primer 2 (Reverse) Con 2 (868-887) 5”-ATTTCGGACCATTTATAACC-3’
The DNA molecular weight marker used was pBR322 DNA/Alu1 Digest containing double stranded DNA segments of 63/57/49, 100/90, 226, 257, 281, 403, 521, 659/656 and 908 bp. The RT-PCR reaction was carried out as per the method of Gouvea et al. 4 with some modifications. The primer mix was prepared by adding viral RNA (2.5 µl), 1.0 µl Forward primer (10 pm), 1.0 µl Reverse primer (10 pm), DEPC treated water (16.0 µl) on ice for 25µl volume in a clean DEPC treated tube. The tube was then placed in thermal cycler (Eppendorf Master Cycler Gradient TM, Germany) at 99°C for five minutes, and snaps chilled on ice. Then 10 X RT buffer (2.5 µl), 100 mM dNTPs (1.0 µl), MMuLV-Reverse Transcriptase 20 U/µl (1µl) were added. This was incubated at 25°C for 10 min. The reverse transcription was carried out at 42°C for 60 min in thermal cycler. This was followed by heat inactivation of MMuLV-RT at 90°C for five minutes. The reaction mixture (25µl) for PCR was prepared by mixing cDNA
(5.0 µl), Forward primer Con 3 (1µl), Reverse Primer Con 2 (1µl) and DEPC treated water (13 µl). The mixtures were heated in thermocycler at 99°C for five minutes and snap chilled on ice. This was followed by the addition of 10X PCR buffer (2.5 µl), 25 mM MgCl2 (1.5 µl), 100 mM dNTPs (0.5µl) and Taq polymerase (3U/µl) 0.5 µl on ice. The amplification was carried out with an initial denaturation at 94°C for five minutes, followed by the sequence of 30 cycles as follows: 94°C for one minute, 50°C for two minutes and 72°C for two minutes. This was followed by final extension at 72°C for 10 min. The PCR product was detected by electrophoresis at 5 V/cm in 1.5 per cent agarose gel in Tris Acetate EDTA buffer (1X).
Agar Gel Immuno Diffusion
Anti-BRV hyper immune serum was raised in rabbits and AGID was done as per the method described by Mohammed et al. 5. The gels were washed in saline overnight and then in distilled water for two hours, dried in a 37°C incubator, and stained with amido black.
Results
Among 124 calves clinically suspected for BRV, 29 (23.39 per cent) were found positive by RNA-PAGE followed by silver staining. It was found that the genome consisting of eleven segments were arranged in clustered pattern of 4:2:3:2 (figure. 1). This pattern is typical for Group A rotavirus. The positive control also showed the same arrangement. All the faecal samples collected from healthy calves and adult animals with diarrhoea were found to be negative.
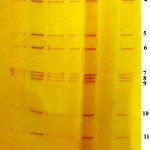 |
Figure 1
|
In the present study, VP4 gene specific RT-PCR was done using the primers Con 2 and Con 3 and an amplicon of 877 bp was generated (Figure. 2). Among 124 samples tested, 35 (28.23 per cent) samples were found positive.
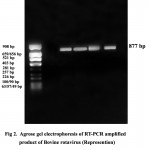 |
Figure 2
|
The presence of BRV antigen in faeces of suspected animals were tested using hyper immune serum raised in rabbit against the reference positive antigen. On viewing the gel against diffuse light, a precipitin line was observed between the reference positive antigen and the central well. The slides were visualized after staining with amido black. Among 124 calves, 16 (12.90 per cent) were found positive by AGID. The formation of a single precipitin line indicated complete identity with the reference antigen. The faecal samples collected from healthy calves and adult animals with diarrhoea were found to be negative.
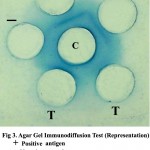 |
Figure 3
|
Age –wise distribution of BRV
The calves suspected for BRV infection were divided into five groups based on age. The results of occurrence of BRV infection among different age groups are presented in Table 1.
Table 1: Age –Wise Distribution of Bovine Rota Virus.
| Age group (weeks) | Samples tested | Samples positive | Percent positive |
| 0 – 2
2 – 4 4 – 6 6 – 8 8 – 10 Total |
33
27 22 20 18 124 |
13
10 7 5 2 35 |
39.39
37.04 31.82 25.00 11.11 28.23 |
The rotaviral infection was more common in first two weeks (39.39 %) followed by two to four weeks (37.04 %) and then four to six weeks (31.82 %). It was less common in six to eight weeks (25.5%) and above (11.11%).
Discussion
The electrophoresis of the extracted RNA in polyacrylamide gels visualized after silver staining revealed the genome consisting of eleven segments. The advantage of RNA-PAGE is that it is possible to identify the group of rotavirus from the clustered arrangement of gene segments. It is convenient to analyze the genomic diversity and heterogeneity of the virus. According to Rodger et al.6, Group A bovine rotavirus showed a migration pattern of 4:2:3:2 in polyacrylamide gels. In the present study, since all the samples showed a similar migration pattern, it can be concluded that Group A rotavirus is more prevalent in the area. In RT- PCR, VP4 gene specific primers were used. The results were in accordance with those obtained by Gentsch et al.7 and Fukai et al.8. Since type specific primers were not used in the study, it was not possible to identify the G and P types of the BRV. The faecal antigen used in AGID was subjected to ultra centrifugation for three hours to obtain concentrated viral antigen. This was used to avoid the presence of multiple lines between the wells.
In this study, RT-PCR detected BRV in six more samples which were found negative in RNA-PAGE. All the RNA-PAGE positive samples were tested as positive by RT-PCR also. These results were in accordance with Arguelles et al. 9. According to them, RT-PCR required 104 particles per milliliter of faeces whereas RNA-PAGE required 1011 particles per millilitre to be detected as positive. Wani et al. 10 also got similar results. Agar Gel Immunodiffusion could detect rotaviral antigen only in 16 (12.90 per cent) samples and hence this test was found to be least sensitive among the three. From the above results, RT-PCR was found to be more sensitive in detecting BRV (Table 2).
Table 2: Comparison of Rna-Page, Rt-Pcr And Agid
|
Test
|
Number of samples tested
|
Number of positive samples
|
Percent positive |
| RNA-PAGE
RT-PCR AGID |
124
124 124 |
29
35 16 |
23.38
28.23 12.90 |
Among the three methods for detecting rotavirus, RT-PCR was found to be the most sensitive (28.23 %) followed by RNA-PAGE (23.38%) and AGID was found to be least sensitive (12.90 %).
The occurrence of infection was highest in the age group below two weeks followed by two to four weeks of age and then four to six weeks of age. It was less common in calves of six to eight weeks and eight to fifteen weeks of age. The age-wise distribution pattern obtained was in accordance with that of Khattar and Pandey 11 who observed that although calves excreting virus were detected upto fifth week of age, maximum number of rotavirus were detected in the first week.. Some other workers12 opined that calves were infected with rotavirus only during the first week of their life. In the present study, 20 adult cows above one year of age with diarrhoea were screened for the presence of BRV, but none of them was found to be positive.
Acknowledgements
We thank the Kerala Agricultural University for funding this study.
References
- Mebus, C.A., Underdahl, N.R., Rhodes, M.B. and Twiehaus, M.J. Uni. Nebraska Agric. Exp. Stat. Res. Bull., 1, 233 (1969).
- Manufacturer’s protocol. Molecular Research Centre, Inc. Cincinnati, OH. (1995).
- Herring, A.J., Inglis, N.F., Ojeh, C.K., Snodgrass, D.R. and Menzies, J.D. J. Clin. Microbiol. 16, 597 (1982).
- Gouvea, V., De castro, L., Timenetsky, D.C., Greenberg, H.B. and Santos, N. J. Clin. Microbiol. 32, 1417 (1994).
- Mohammed, K.A., Babiuk, L.K., Saunders, J.R. and Acres, S.D. Vet. Microbiol. 3, 168 (1978).
- Rodger, S.M., Schnagl, R.D. and Holmes, I.H. J. Virol. 24, 421 (1977).
- Gentsch, J.R., Glass, R.I., Woods, P., Gouvea, V., Gorziglia, M. and Flores, J. J. Clin. Microbiol. 30, 1620 (1992).
- Fukai, K., Sakai, T. and Kamata, H. Aust. Vet. J. 76, 440(1998).
- Arguelles, M.H., Villegas, G.A., Castello, A., Abrami, A., Ghiringhelli, P.D., Semorile, L. and Glikmann, G. J. Clin. Microbiol. 38, 475 (2000).
- Wani, S.A., Bhat, M.A., Ishaq, S.M. and Quereshi, S. Indian Vet. J. 83, 707 (2005).
- Khattar, S. and Pandey, R. Indian J. Virol. 2, 230 (1986).
- Tzipori, S.R., Makin, T.J., Smith, M.L. and Krautil, F.L. J. Clin. Microbiol. 13, 1125 (1981).

This work is licensed under a Creative Commons Attribution 4.0 International License.





