Manuscript accepted on : 23-04-2020
Published online on: 08-05-2020
Plagiarism Check: Yes
Reviewed by: Ayush Dogra ![]()
![]()
Second Review by: Revathi Shenoy ![]()
![]()
Gut Microbial Communities of Adult Honey Bee Workers (Apis Mellifera)
Marfat Alatawy1,2* , Sanaa G. Al-Attas1
, Sanaa G. Al-Attas1
![]() , Ahmad I. Assagaf1
, Ahmad I. Assagaf1 , Abdullah Al-Shehri3
, Abdullah Al-Shehri3
![]() , Khalid M. Alghamdi1
, Khalid M. Alghamdi1 , Ahmed Bahieldin1,4
, Ahmed Bahieldin1,4
1Department of Biological Sciences, Faculty of Science, King Abdulaziz University, Jeddah, Saudi Arabia.71491
2Department of Biology, College of Science, Tabuk University, Tabuk, Saudi Arabia.74191
3Department of Arid land Agriculture, Faculty of Metrology, Environment and Arid land Culture, King Abdulaziz University, Jeddah, Saudi Arabia.80200
4Department of Genetics, Faculty of Agriculture, Ain Shams University, Cairo, Egypt. 11241
Corresponding Author E-mail : mevalatawi@ut.edu.sa
DOI : http://dx.doi.org/10.13005/bbra/2838
ABSTRACT: Apis mellifera honey bees are highly valued insects due to their roles in honey production and pollinating some globally important crops. However, honey bee colonies have been decreasing significantly around the world and this has drawn the attention to investigate factors that can affects bees health such as gut microbiome. Gut microbiome is considered an essential part of a honey bee system. Honey bees gut microbiome consists of a nine core species which mostly obtained by social transmission. Current findings on gut microbiome specific strain variations, results on their metabolic and nutritional roles, and links between gut microbial disruption and disease states, have drawn the attention to how microbiota impacts bee health, and also being a potential model to study ecology as well as gut symbionts development. Overall, roles of gut microbiome in honey bees development are becoming much more evident.
KEYWORDS: Apis Mallifera; Dysbiosis; Gut Microbiome; Symbionts
Download this article as:| Copy the following to cite this article: Alatawy M, Al-Attas S. G, Assagaf A. I, Al-Shehri A, Alghamdi K. M, Bahieldin A. Gut Microbial Communities of Adult Honey Bee Workers (Apis Mellifera). Biosci Biotech Res Asia 2020;17(2). |
| Copy the following to cite this URL: Alatawy M, Al-Attas S. G, Assagaf A. I, Al-Shehri A, Alghamdi K. M, Bahieldin A. Gut Microbial Communities of Adult Honey Bee Workers (Apis Mellifera). Biosci Biotech Res Asia 2020;17(2). Available from: https://bit.ly/2WHIofw |
Introduction
Honey bees, such as Apis mellifera, are pollinators for many important crops and they are widely domesticated for their honey production. Therefore, honey bees are essential for our food supply. However, Beekeeping industry all over the world is suffering from huge economic losses. Since 20061, there was a significant reduction in the honey bee colonies. This decline is influenced by multiple factors such as environmental stresses, pollution, exposure to pesticides or antibiotics, as well as foreign pathogens. This decline has drawn attention towards understanding the microbial relations with these species, both symbiotic and pathogenic relations. One way to overcome this problem is to improve bee health by investigating the microbial diversity in bees and its impact on the host1. In this field, the terms ‘microbiome’ and ‘microbiota’ are used interchangeably to represent the microbial community living within a larger host system or in any compact environment. However, microbiome is usually used in science for the collective genetic material of such microbiota. This is the concept that will be used in this review analysis, since microbiome characterisation techniques are primary steps in microbiota characterisation2.
The gut microbiome highly impacts bees’ health as it are involved in metabolism, nutrient absorption, immunity and development. Since honey bee gut has a relatively simple microbial composition and experimental amenability, they are considered a promising organism to investigate the essential aspects of gut microbiology3. The majority of the bacterial 16S rRNA sequences (95%) detected for the adult honey bee belongs to phyla Firmicutes, Actinobacteria and Proteobacteria (Alpha proteobacteria, Betaproteobacteria, Gamma proteobacteria), and therefore, are considered the core species of the gut microbial content in honey bees. In addition, there is a high level of diversity in gut bacterial species, with different compositions, which could be associated with the nutritional and health status of the honey bee4.
The microbial community within honey bees has been studied and analyzed using culture-based techniques5. However, with the advancement in molecular tools, investigating the microbial composition and structure of the honey bee’s gut became much easier. Numerous publications were published concerning this research, though, their findings needs refining and clarification of such point for further reviewing and meta-analysis. This review aims to summarize results regarding the structure and transmission of microbiome in adult honey bees guts. In addition, focuses on the potential roles of the core bacterial species; the essential structure of the normal gut microflora.
Honey Bees (Apis Mellifera)
Honey bee, or Apis mellifera, belongs to the order Hymenoptera and the superfamily Apoidea, and it is that is highly regarded due to its importance to human health and ecosystems (Table 1). For instance, in addition to honey production, the honey bee plays a major role in the pollination process of different economically important crops6. Without these pollinators, the yields of some seed, fruit and nut crops have decreased by more than 90%7,8.
Honey bees live in a eusocial system of perennial colonies with overlapping generations, a reproductive section of labour and a brood care division. In addition, each colony is composed of three castes7 as the following: the female worker bees, which fluctuate in number between 15,000 in the winter and 50,000 in the summer, the male drones, which usually exist only in the spring and have numbers in the few hundreds, and the female reproductive queen bee.
Worker bees are further classified based on age, as they have different roles in the hive depending on their ages. Firstly, Younger bees, which feed on the lipid and protein contents of processed pollens (bee bread), are normally constrained to the hive and are involved in the rearing of the brood. For this reason, they are called ‘nursing bees’. Secondly, Older bees ‘‘foragers’’, are tasked with looking for nectar and pollen outside of the hive. Food is then brought into the hive and passed from bee to bee by an exchange process called trophallaxis, which turns the honey bee bread into food products9.
In recent decades, the number of honeybee colonies has decreased dramatically throughout the world10. The International Cost Action FA0803 COLOSS (Colony LOSS) network which developed by the European Union, is an association of 161 members from more than 40 different countries that was established to address and prevent global honeybee colony collapse worldwide. A monitoring study carried out by the COLOSS team during the winters of 2007-2008 reported honeybee losses of in the USA about 30%, 25% in Japan, 1.8%-53% in Europe, and 10%-85% in the Middle East11. These losses were influenced by multiple factors, such as environmental stress conditions, lack of nutrition (nectar and pollen), extensive use of insecticides, and biotic stresses such as infection by pathogenic parasites (e.g. Acarapis woodi, Varroa destructor, Tropilaelaps spp., microsporidia Nosema spp.), pathogenic fungi (e.g. Ascosphaera apis) or bacteria (e.g. Paenibacillus larvae, Melissococcus plutonius), in addition to more than 18 different viruses, including deformed wing virus (DWV)12.
Table 1: classification of Apis melifera
| Taxa | Names |
| kingdom | Animalia |
| Phylum | Arthropoda |
| Class | Insecta |
| Order | Hymenoptera |
| Family | Apiidae |
| Genus | Apis |
| Species | Apis melifera |
The Honey Bee as a Model System for Gut Microbiota Research
As previously mentioned, the honeybee system model is promising for investigating gut microbiota and understanding essential aspects of gut microbiology because of its relatively simple microbial composition and experimental amenability. In addition, honey bees can serve as microbiota-free hosts, which enable the researchers to investigate how microbiotas relate to the host’s phenotype (e.g. conditions and diseases) more accurately. In mammals, it is only possible to obtain microbiota-free individuals through Caesarean sections, and then the mammals must be kept in special housings. However, a germ-free insect can be produced by chemical sterilisation of the egg surface, nevertheless, the use of antibiotics to obtain large numbers of microbiota-free hosts interferes with, and threatens the normal honeybee life cycle.
The honeybee microbiome shares many features with the human microbiome, which other insects do not, such as the following: At first, some bacterial species have adapted to, and dominated the host’s gut microbiome and usually are not seen elsewhere. In addition, microbiome is highly important element in overall health of both systems. Moreover, the strains of the local bacterial species are highly diverse. Furthermore, when exposed to antibiotics or chemicals, the continuous use of antibiotics has impacted the microbial diversity within the human gut, resulting in high rates of resistance factors. Similarly, antibiotic use has interfered with honeybee gut communities3.
The Gastrointestinal Tract of Worker Honey Bees
The digestive tract of the honey bee is divided into three different compartments: foregut, midgut and hindgut. The foregut contains the crop, the midgut is the middle part of the tract and the hindgut includes the ileum and rectum (Figure 1). Different areas of the digestive system provide different environments for the bacterial symbionts, which have different roles in essential biological activities (e.g. metabolism and absorption). Food collected by the foraging workers is transported through the oesophagus into the crop. The crop is a muscle-lined organ that has the ability to expand for large quantities of nectar. Accordingly, it is sometimes called the ‘honey stomach’. Although the crop can carry large amounts of nectar and nutrients that the microbes utilise for energy, it accommodates only small amounts of bacteria. In the process of honey production, the crop is continuously filled with and emptied of nectar. This can compromise the integrity of the gut microbiome and inhibit bacterial colonisation. In addition, the crop produces different enzymes to process nectar. It has been hypothesized that these enzymes are responsible for the antimicrobial activity of the honey, as they prevent bacterial growth within the crop13. Then, food and nutrients are moved into the midgut through the proventriculus, a muscular tissue located under the crop, which contains valves and can protect the midgut against foreign particles. The midgut is the biggest compartment of the gastrointestinal (GI) tract in which most of the food digestion and absorption occur. This is why it is also called the ventriculus, or ‘true stomach’. The epithelial layer of the midgut contains different enzymes that can metabolize protein, fat and sugar. Anything that remains after digestion is then moved to the ileum through the pylorus, an interceptive valve between the midgut and the ileum. Lastly, in the hindgut, the ileum is a smaller part of the GI tract between the midgut and rectum that has deep infoldings to provide a large surface area to collect nutrients that were not absorbed in the midgut. The rectum is the distal part of the GI tract, and similar with the crop, it has the ability to distend to fit waste products after food digestion. This allows the workers to retain waste materials so they can dispose them through defecation outside of the hive. The rectum is a relatively stationary environment, and its waste products (mostly empty pollen exines) can provide a good source of nutrition for bacteria, as the carbohydrates found in the exine layer are recalcitrant to the honey bee’s digestive enzymes14.
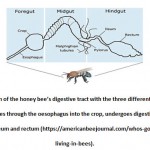 |
Figure 1: Illustration of the honey bee’s digestive tract with the three different |
Honeybee Gut Microbiome
Because honey bees are social insects that have close relations within their community and have large colonies, they can provide unique ways for bacterial microbiome nutrition9. As previously mentioned, the honeybee gut microbiome is essential to the host’s entire system and plays a major role in metabolism, food absorption, immunity and development15. The majority bacterial sequences detected for the adult honey bee consists of eight characteristics bacterial phylotypes (Table 2). They are two alpha proteobacteria, Alpha1 and Alpha2, of Acetobacteraceae; two gamma proteobacteria, Gamma 1, recently identified as Gilliamella apicola16, and Gamma 2, recently identified as Frischella perrar17; two lactobacillus, Firm-4 and Firm-5; one betaproteobacteria, which identified as Snodgrassella alvi16; with one Bifidobacterium, identified as Bifido. While many bacterial phylotypes of the honeybee gut are closely related to those found in other insects, three specific phylotypes (G. apicola, F. perrara and S. alvi) have only been found in honey bees and bumble bees, up to date1.
Although the identified core microbiotas are comprised of few phylotypes, their underlying species have shown relatively high strain variations. Two species in particular have demonstrated high strain variations. They are G. apicola (belonging to gamma proteobacteria: Orbales) and S. alvi (belonging to Betaproteobacteria: Nesseriales)18. This has also been observed with Lactobacilli and Bifidobacterium spp., which are associated with honey bees19. Despite the fact that there are different honey bees, colonies and distributions around the world, it has been suggested that this coherent structure of the same phylotypes plays a major role in the health of all bees. Moreover, the strain variation within the different phylotypes may offer different functionalities and could also play a major role in bees’ health18.
Studies conducted on colonization trends and the bacterial composition of the honeybee GI tract reveals that there is a lack of bacteria until the honey bees reach the age of 4-6 days. Young workers can be inoculated in different ways, including environmental factors (via bee bread and comb) and also through interactions with older bees within the colony. Once they are inoculated, different gut compartments will have different bacterial communities; for example, the crop and midgut are occupied by very few bacteria (104 and 106, respectively). In contrast, the hindgut, which made up of the ileum and rectum, harbours large communities with distinct compositional profiles. The ileum and rectum have total bacterial amounts of around 107 and 108, respectively14.
Table 2: The core gut microbiota and its distribution41, 42
| Taxonomic classification of core gut microbiota for adult Apis mellifera workers | ||||||
| Phylotype | Bacteria phylum | Order | Family | Genus | Species | Primary locations |
| Gamma1 | Gamma proteobacteria | Orbales | Orbaceae | Gilliamella | apicola | Adult midgut, hindgut (ileum) |
| Gamma2 | Gamma proteobacteria | Orbales | Orbaceae | Frischella | perrara | Adult hindgut (ileum) |
| Beta | Betaproteobacteria | Neisseriales | Neisseriaceae | Snodgrassella | alvi | Adult hindgut (ileum) |
| Firm4 | Firmicutes | lactobacillales | Lactobacillaceae | Lactobacillus | mellifer
|
Adult hindgut (rectum) |
| Firm5 | Firmicutes | lactobacillales | Lactobacillaceae | Lactobacillus | apis
|
Adult hindgut (ileum, rectum) |
| Bifido | Actinobacteria | Bifidobacteriales | Bifidobacteriaceae | Bifidobacterium | asteroides
|
Adult hindgut (rectum) |
| Alpha1
|
Alpha proteobacteria | Rhizobiales | Bartonellaceae
|
Bartonella
|
apis
|
Adult gut
|
| Alpha2 | Alpha proteobacteria | Rhodospirillales | Acetobacteraceae | Acetobacter | aceti | Larval gut, adult crop, nectar, honey, hive, adult hindgut |
Honeybee Microbiota Transmission
Honey bees share their gut bacteria with other members of the colony through oral-faecal routes. This occurs through trophallactic interactions, consumption of the stored pollen and bee bread, contact with older bees in the hive, and exposure to hive materials during the early-adult stage20, 21. A number of studies showed a slight difference in microbial gut composition due to factors such as host age, diet, caste, seasonal or geographical alterations. These factors were shown to be influential to the bee microbiome. In fact, a lack of nutrition leads to gut microbiome disruption, resulting in high disease and mortality rates22.
Furthermore, honey bees are highly social insects within the hive, and this is the key element in the transmission of microbiota between hosts (Figure 2). After pupating in capped cells within the colony, adult honey bees with germ-free guts emerge21,23. The honey bees’ gut was manually extracted just after they were removed from honeycomb cells and were kept in sterile conditions, but they did not show any significant microbial composition. This was validated using qPCR with broadly used primers. In this study, Kwong et al. 20, 21 found that newly emerging bees, after freely leaving honeycomb cells, can be inoculated on the frame surface with residual gut symbionts.
In another study, the developmental stages of gut microbiota were characterized by testing marked cohorts of colonial workers, using 16S rDNA amplicons to assess microbial size and composition of different areas of the gut24. Initially, the microbial composition was relatively small and random and environmental species were predominant with no significant difference between microbial compositions in different areas of the gut. Around three days later, microbial compositions were found to be about >107 bacteria. These are predominantly characteristic species of a conventional bee’s gut. The ileum and rectum also started to show ‘ordinary’ bacterial compositions. By the eighth day, this microbial concentration plateaued around 109. It has also been found that when established, gut microbial populations are usually stable throughout a worker bee’s transitional states25. This shifting from an initial chaotic microbial community to one dominated by ‘adult’ bacteria, greatly mimics human infant’s gut microbiota growth26.
Since a sustained microbial community is established before worker bees emerge from the hive, it can be determined that bacterial transmissions occur in between nestmates or through hive components, such as wax surfaces. In the laboratory, in order to obtain typical gut microbiome development, this was highly achieved through interacting younger bees with older ones, or fed their macerated hindguts23. In studies on potential routes of transmission, researchers found that oral trophallaxis, a common way for bees to communicate and exchange food, was not essential for bacterial transmission. These results are consistent with other results which stated that foregut regions are occupied by a limited number of bacteria15. Although many members of the gut microbiome can be obtained through interacting with different hive components that had been previously contacted by other hive bees, a faecal route remains important, especially for Snodgrassella alvi, Gilliamella apicola and Frischella perrara27. On the other hand, species of the Acetobacteraceae family could be transmitted through pollens that are stored for nutrition10.
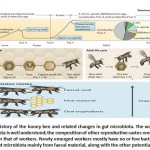 |
Figure 2: Life history of the honey bee and related changes in gut microbiota |
Metabolic Characterisation
Meta-transcriptomic datasets and metagenomic and total genomic sequencing for many members of honey bees gut microbiota have provided insight into the lifestyles and prospective roles of gut microbiota19, 20, 21, 27, 28, 29. As in the human gut, microbiota are involved in the metabolism of carbohydrates in the bee gut. In honey bees, this is carried out by bee-specific species, Lactobacillus and Bifidobacterium, which are genera that have members in mammalian guts.
Experimental tests on the B. asteroids species cluster in bees have identified highly abundant and diverse genes involved in carbohydrate use, but they were not found in both of its relatives or in other microbial community members30. In terms of phylogenetics, Bifidobacterium spp. associated with bees are branches of the same major groups of Bifidobacterium spp. found in mammals and, therefore, this exhibits a great parallel model system to study microbiota adaptation in human as well 31. Indeed, bee-derived B. asteroids have shown clear differences from those in mammals, such as their capacity for aerobic respiration4, which may indicate specific conditions within the bee’s gut (e.g. different oxic conditions). On the other hand, strains of Lactobacillus in honey bees were found to be different from those in mammals, and in general, the strains found in bees and mammals fall under two separate phylogenetic clades, Firm-4 and Firm-5, but also rare under a third clade, Firm-327. Current genome sequencing of Lactobacillus Firm-4 and Firm-5 identified multiple phospho-transferase enzyme systems essential for sugar uptake in both, but mainly in Lactobacillus Firm-5.
In addition, both Lactobacillus and Bifidobacterium strains in bees carries large proteins on their cell-surface with putative structure. Their functions are unknown, but it is suggested that the proteins have roles in adhesion or degradation of plant materials10. Bees also harbour gene clusters of B. asteroids and Lactobacillus Firm-5 that are responsible for the production and utilization of trehalose, which is a disaccharide molecule used as an energy reservoir in insects. On the other hand, mammals use glycogen for energy storage. Moreover, in mammals, Lactobacillus spp. and Bifidobacterium spp. normally carries genes responsible for biosynthesis and degradation of glycogen. However, these genes are not present in bees19.
Gilliamella apicola is another dominant fermentative bacteria in honey bees’ guts. It belongs to the bacterial order Orbales, which is mostly linked to insects16. Genomic analysis has identified a wide range of genes in G. apicola strains involved in the uptake and fermentation of sugars, but it has also identified an incomplete tricarboxylic acid cycle and a degenerate aerobic respiratory chain20,21. Another two microbial members found in bees and associated with Orbales are Schmidhempelia bombi and Frischella perrara, which have similar activities but are also carbohydrate fermenters17, 32,33.
Since bees thrive on nectar, honey and pollen, which are high in carbohydrates, it is expected that their microbiota includes members that can process these types of foods. They do indeed have F. perrara,G. apicola, Lactobacillus Firm-4, Lactobacillus Firm-5 and bee-associated B. asteroids, which have the ability to metabolise glucose and fructose, the main sugars types of bee’s diet. Some other strains carry genes capable for utilization of other rare sugars such as arabinose, mannose, raffinose, lactose and galactose, which are indigestible by bees and potentially toxic15. Different microbial species lead to different end products of the fermentation process, but usually, their end products are lactic acid and acetate. DNA and RNA Metagenomic sequencing analysis have revealed high levels of expression of those genes involved in fermentation27. This was also verified using culture-based assays10.
The last core member of the honey bees’ microbiota is S. alvi, an obligate microaerophilic bacterium that belongs to the Neisseriaceae family. The distribution of S. alvi in the peripheral areas of the gut lumen is consistent with their dependence on aerobic respiration, where the epithelial surface of insects’ guts has the maximum oxygen concentrations35. Remarkably, S. alvi no longer relies on carbohydrate glycolysis for the production of energy, but rather, it depends on aerobic oxidation of carboxylates, such as acetate, malate, citrate and lactic acid. Since different resources can be used for energy, S. alvi can coexist with other fermentative bacteria in the same gut environment. Besides, this metabolic difference is indicative for the syntrophic interaction, as many carbohydrates fermentation products (acetate, formate and lactic acid) are substrates used by S. alvi 16, 20, 21.
Parasaccharibacter apium is another bacterium that has a specific niche. Although rare in adult worker bees’ guts, P. apium appears to be abundant within the hive food stores, larvae’s and queens’ guts. Remarkably, P. apium is able to thrive on royal jelly, which is a toxic environment to most other bacteria, and is found in royal jelly-producing glands of worker bees. Although P. apium is an Acetobacteraceae member, genomic data from P. apium26 and other closely related phenotypes indicated that P. apium do not produce acetic acid through sugar and alcohol oxidation. On the other hand, it looks to be well suited to the aerobic, acidic and high-sugar conditions found in royal jelly, nectar and honey37.
Role of the Bee Gut Microbiome in Health, Nutrition and Protection against Pathogens
The commensal microflora bacteria are potential symbionts. These bacterial symbionts evolve with their host15, and many bacteria have been identified to have a protective role against foreign pathogens27. In addition, different symbionts have unique roles within the gut, and their specific positioning is important for the functional integrity of the gut10. Hamdi et al. 12 showed that gut symbionts are significant for bee health, and dysbiosis within the gut microbiota can lead to diseases. In fact, a lack of nutrition leads to disruption of the gut microbiome, resulting in high disease and mortality rates. This disruption of the microbiome (dysbiosis) affects the development of adult worker bees through inhibition of important gene expression, such as vitellogenin. Vitellogenin is a phospholipoglyco-protein that affects multiple aspects of the honeybee life cycle. It is a female-specific egg yolk protein with an essential function related to oogenesis4, 38, 39. Moreover, dysbiosis may cause bees to be unable to adapt to stressful conditions, such as heat or poor nutrition, and in turn, these conditions can impact the microbiome.
The consistency in the microbiota system of honey bees has led to hypotheses about its potential role in honey bee lives, whether symbiotic or not. Different bacterial groups have been shown to produce short-chain fatty acids, such as acetic acid or lactic acid (e.g. Lactobacilli, Bifidobacteria, Acetobacteraceae and Simonsiella), which honey bees consume as a food supplement. Moreover, gut bacteria makes it possible for honey bees to degrade pollen, which is coated with exine layers that are resistant to most digestive enzymes, in order to use the intine for nutrition. Gut bacteria species of honey bees show a high level of diversity and have different compositions, which could be associated with the nutritional and health status of the host15.
Genomic and metabolic analyses of core species in the bee gut (Lactobacillus, Bifidobacterium, and G. apicola) indicate that these bacteria are able to utilize different groups of plant-origin carbohydrates. In a comparative study of microbiota-free bees and bees with a conventional gut microbiome, the gut microbiome was shown to have many physiological effects. The study showed a major positive effect on gut size, weight gain, insulin and vitellogenin signalling and sucrose sensitivity27. These physiological changes may impact bees’ immunity, stress tolerance and overall health. In addition, the gut microbiome has been proven to be a cofactor in fighting infections in both honey bees and bumble bees. In two different studies, the faecal matter of wild-type worker bees was inoculated with a microbiota-free B. terrestis, which showed immunity against the trypanosomatid gut parasite Crithidia bombi in contrast to other workers that were not inoculated. Moreover, the transplanted bacteria’s ability to protect against infections was dependent upon their source rather than the bees’ original colonies, indicating that the level of protection was dependant on the different microbiota compositions10. Two symbionts of the honey bee, S. alvi and G. apicola, were found to be enriched in gene encoding for biofilm formation. When these were viewed using fluorescence microscopy, it appeared that the epithelium layer of the host ileum was enveloped by the two species, suggesting protective characteristics related to biofilm functions, such as providing a protective layer against parasites4, 38, 39.
Conclusion
Honey bees are highly valued insects throughout the world, owing to their role in honey production and pollinating many globally important crops. However, their populations have recently declined and this drew attention to the potential factors affecting their health, such as microbiota 4, 38, 39. Bee guts occupies a unique and stable microbiome, therefore, it is an essential part of bee biology. In addition, current experimental studies have revealed a crucial roles of the gut microbiome in bees health such as nutrition and immunity. Understanding this microbial community provides visions into how to improve bee health, and into overall unresolved aspects of host-microorganism symbiosis. This consequently helps to overcome the reductions in bee populations around the world and maintains food security.
Acknowledgments
The authors would like to acknowledge Prof. Dr. Rashad R. Al-Hindi, Professor of food Microbiology and Head of Microbiology program, Department of Biological Sciences, Faculty of Science, King Abdulaziz University, Jeddah, Saudi Arabia, for revising of the final version of the manuscript.
Conflict of interest
The authors declare that there is no conflict of interest
Funding Source
There are no funding resources for this article.
References
- Moran N. A., Hansen A. K., Powell J. E., Sabree Z. L. Distinctive gut microbiota of honey bees assessed using deep sampling from individual worker bees. PloS One. 2012;7(4):e36393.
- Romero S., Nastasa A., Chapman A., Kwong W. K., Foster L. J. The honey bee gut microbiota: strategies for study and characterization. Insect Biol. 2019:28(4):455-472
- Bonilla-Rosso G. and Engel P. Functional roles and metabolic niches in the honey bee gut microbiota, Opin. Microbiol. 2018a;43:69-76.
- Crotti E., Sansonno L., Prosdocimi E. M., Vacchini V., Hamdi C., Cherif A., Gonella E., Marzorati M., Balloi A. Microbial symbionts of honeybees: A promising tool to improve honeybee health. Biotechnol. 2013;30(6):716–722.
- Gilliam M., Roubik D. W. and Lorenz, B. J. Microorganisms associated with pollen, honey, and brood provisions in the nest of a stingless bee, Melipona fasciata. Apidologie. 1990;21(2):89–97.
- Khan K. A., Ansari M. J., Al-Ghamdi A., Nuru A., Harakeh S., Iqbal J. Investigation of gut microbial communities associated with indigenous honey bee (Apis mellifera jemenitica) from two different eco-regions of Saudi Arabia. Saudi J Biol Sci. 2017;24(5):1061–1068.
- Winston M. L. (ed): The Biology of the Honey Bee. 2nd Harvard University Press. 1990; pp 197-199
- Klein A. M., Vaissière B. E., Cane J. H., Steffan-Dewenter. I., Cunningham S. A., Kremen C., Tscharntke T. Importance of pollinators in changing landscapes for world crops. Proc Biol Sci. 2007;274(1608):303–313.
- Rokop Z. P., Horton M. A. and Newton I. L. G. Interactions between cooccurring lactic acid bacteria in honey bee hives. Environ. Microbiol. 2015;81(20):7261–7270.
- Yun J. H., Jung M. J., Kim P. S., Bae J. W. Social status shapes the bacterial and fungal gut communities of the honey bee. Sci Rep. 2018;8(1):1–12.
- Carreck N. and Neumann P. Honey bee colony losses. Apicul. Res. 2010;49(1):1.
- Hamdi C., Balloi A., Essanaa J., Crotti E., Gonella E., Raddadi N., Ricci I., Boudabous A., Borin S., Manino A., Bandi C., Alma A., Daffonchio D., Cherif A. Gut microbiome dysbiosis and honeybee health. Appl. Entomol. 2011;135(7):524–533.
- Anderson K. E., Sheehan T. H., Mott B. M., Maes P., Snyder L., Schwan M. R., Walton A., Jones B. M., Corby-Harris V. Microbial ecology of the hive and pollination landscape: Bacterial associates from floral nectar, the alimentary tract and stored food of honey bees (Apis mellifera),” PloS One. 2013;8(12):e83125.
- Martinson V. G., Moy J. and Moran N. A. Establishment of characteristic gut bacteria during development of the honeybee worker. Environ. Microbiol. 2012;78(8):2830–2840.
- Zheng H., Powell J. E., Steele M. I., Dietrich C., Moran N. A. Honeybee gut microbiota promotes host weight gain via bacterial metabolism and hormonal signaling. Proc Natl Acad Sci U S A. 2017;114(18):4775–4780.
- Kwong W. K. and Moran N. A. Cultivation and characterization of the gut symbionts of honey bees and bumble bees: description of Snodgrassella alvi gen. nov., sp. nov., a member of the family Neisseriaceae of the Betaproteobacteria, and Gilliamella apicola gen. nov., sp. nov., a member of Orbaceae fam. nov., Orbales ord. nov., a sister taxon to the order ‘Enterobacteriales’ of the Gammaproteobacteria. Int J Syst Evol Microbiol. 2013;63(6):2008–2018.
- Engel P., Kwong W. K. and Moran N. A. Frischella perrara gen. nov., sp. nov., a gammaproteobacterium isolated from the gut of the honeybee, Apis mellifera,” Int J Syst Evol Microbiol. 2013;63(10):3646–3651.
- Engel P., Stepanauskas R. and Moran, N. A. Hidden diversity in honey bee gut symbionts detected by single-cell genomics,” PLoS Genet. 2014;10(9):e1004596.
- Ellegaard K. M., Tamarit D., Javelind E., Olofsson T. C., Andersson S. G., Vásquez A. Extensive intra-phylotype diversity in lactobacilli and bifidobacteria from the honeybee gut. BMC Genomics. 2015;16(1):284.
- Kwong W. K., Engel P., Koch H., Moran N. A. Genomics and host specialization of honey bee and bumble bee gut symbionts. Proc Natl Acad Sci U S A. 2014;111(31):11509–11514.
- W. K., Mancenido. A. L. and Moran, N. A. Genome sequences of Lactobacillus sp. strains wkB8 and wkB10, members of the Firm-5 clade, from honey bee guts. Genome Announc. Am 2014;2(6):e01176-14.
- Bonilla-Rosso G. and Engel P. Functional roles and metabolic niches in the honey bee gut microbiota. Opin. Microbiol. 2018b;43: 69–76.
- Powell J. E., Martinson V. G., Urban-Mead K., Moran N. A. Routes of acquisition of the gut microbiota of the honey bee Apis mellifera. Environ. Microbiol. Am Soc Microbiol, 2014;80(23):7378–7387.
- Vásquez A., Forsgren E., Fries I., Paxton R. J., Flaberg E., Szekely L., Olofsson T. C. Symbionts as major modulators of insect health: Lactic acid bacteria and honeybees. PLoS One. 2012;7(3):e33188.
- Kapheim K. M., Rao V. D., Yeoman C. J., Wilson B. A., White B. A., Goldenfeld N., Robinson G. E. “Caste-specific differences in hindgut microbial communities of honey bees (Apis mellifera),” PloS One.. 2015;10(4):e0123911.
- Amdam G. V., Fennern E. and Havukainen H. Vitellogenin in honey bee behavior and lifespan. Honeybee Neurobiology and Behavior. 2012;doi: 10.1007/978-94-007-2099-2_2.
- Raymann K. and Moran N. A. The role of the gut microbiome in health and disease of adult honey bee workers. Curr Opin Insect Sci. 2018;26: 97–104.
- Engel P., Martinson V. G. and Moran N. A. Functional diversity within the simple gut microbiota of the honey bee. Proc Natl Acad Sci U S A. 2012;109(27):11002–11007.
- Lee F. J., Rusch D. B., Stewart F. J., Mattila H. R., Newton I. L. Saccharide breakdown and fermentation by the honey bee gut microbiome. Environ Microbiol. 2015;17(3):796–815.
- Bottacini F., Milani C., Turroni F., Sánchez B., Foroni E., Duranti S., Serafini F., Viappiani A., Strati F., Ferrarini A., Delledonne M., Henrissat B., Coutinho P., Fitzgerald G. F., Margolles A., van Sinderen D., Ventura M. Bifidobacterium asteroides PRL2011 genome analysis reveals clues for colonization of the insect gut. PLoS One. 2012;7(9):e44229.
- Lugli G0 A., Milani C., Turroni F., Duranti S., Ferrario C., Viappiani A., Mancabelli L., Mangifesta M., Taminiau B., Delcenserie V., van Sinderen D., Ventura M. Investigation of the evolutionary development of the genus Bifidobacterium by comparative genomics. Environ. Microbiol. 2014;80(20): 6383–6394.
- Martinson V. G., Magoc T., Koch H., Salzberg S. L., Moran N. A. Genomic features of a bumble bee symbiont reflect its host environment. Environ. Microbiol. 2014;80(13):3793–3803.
- Engel P., Vizcaino M. I. and Crawford J. M. Gut symbionts from distinct hosts exhibit genotoxic activity via divergent colibactin biosynthesis pathways. Environ. Microbiol. 2015;81(4):1502–1512.
- Olofsson T. C., Alsterfjord M., Nilson B., Butler E., Vásquez A. Lactobacillus apinorum sp. nov., Lactobacillus mellifer sp. nov., Lactobacillus mellis sp. nov., Lactobacillus melliventris sp. nov., Lactobacillus kimbladii sp. nov., Lactobacillus helsingborgensis sp. nov. and Lactobacillus kullabergensis sp. nov., isol. Int J Syst Evol Microbiol. 2014;64(Pt 9):3109.
- Brune, A. Symbiotic digestion of lignocellulose in termite guts. Rev. Microbiol. 2014;12(3):168-180.
- Corby-Harris V., Snyder L. A., Schwan M. R., Maes P., McFrederick Q. S., Anderson K. E. Origin and effect of Alpha 2.2 Acetobacteraceae in honey bee larvae and description of parasaccharibacter apium gen. nov., sp. nov. Environ. Microbiol. 2014;80(24):7460–7472.
- Chouaia B., Gaiarsa S., Crotti E., Comandatore F., Degli Esposti M., Ricci I., Alma A., Favia G., Bandi C., Daffonchio D. Acetic acid bacteria genomes reveal functional traits for adaptation to life in insect guts. Genome Biol. Evol. 2014; 6(4):912–920.
- Cox-Foster D. L., Conlan S., Holmes E. C., Palacios G. , Evans J. D., Moran N. A., Quan P. L., Briese T., Hornig M., Geiser D. M., Martinson V., vanEngelsdorp D., Kalkstein A. L., Drysdale A., Hui J., Zhai J., Cui L., Hutchison S. K., Simons J. F., Egholm M., Pettis J. S., Lipkin W. I. A metagenomic survey of microbes in honey bee colony collapse disorder. Science. 2007;318(5848):283–287.
- Fürst M. A. McMahon D. P., Osborne J. L., Paxton R. J., Brown M. J. Disease associations between honeybees and bumblebees as a threat to wild pollinators. Nature. 2014;506(7488):364-366.
- Moran N. A. Symbiosis. Current Biology. 2006;16(20):R866–R871.
- Moran N. A. Genomics of the honey bee microbiome. Opin. Insect Sci. 2015;10:22–28.
- Kwong W. K. and Moran N. A. Gut microbial communities of social bees. Rev. Microbiol. 2016;14(6):374–384.

This work is licensed under a Creative Commons Attribution 4.0 International License.





