Manuscript accepted on : 25 January 2017
Published online on: --
Plagiarism Check: Yes
Concomitant Ability of Siderophore Against iron Paucity and Fusarium wilt in Lycopersicon Esculentum
Stuti Sah1, Manvika Sahgal2 and Rajni Singh1
1Amity Institute of Microbial Biotechnology, Amity University, Sector 125, Noida, UP, India.
2Department of Microbiology, CBSH, G. B. Pant University of Agriculture and Technology, Pantnagar, India.
Corresponding Author E-mail: rsingh3@amity.edu
DOI : http://dx.doi.org/10.13005/bbra/2449
ABSTRACT: Insoluble iron present in soil severely restricts its bioavailability for plant growth. Microorganisms present in the rhizosphere release siderophore to make it available to the plants. Among others, fluorescent pseudomonads are known to exert extensive biocontrol action against soil and root borne phytopathogens through release of siderophores. A total of 172 rhizobacterial isolates were obtained from two different ecosystems viz. forests and agricultural soils, among these 34 were found to produce siderophore with an apparent decrease in siderophore production when supplemented with 20 µM iron. With the aim of utilizing siderophore production as an antagonist against Fusarium oxysporium, isolates, four Pseudomonas isolates namely RSP7 (KR051487), RSP8 (KR051488), RSP3 (KR051489) and RSP5 (KR051490) were selected. Paired t-test analysis resulted in showing antagonism of RSP5 as 48.5% on plate, and the paired t-test value as 14with a significance of P< 0.01. RSP showed an antagonism of 20% on plate with t value of 31.1 and P< 0.01. Paired t-test analysis proved a highly significant antagonism with isolate RSP7 (t = 37.37, P < 0.001). The results among the four isolates are comparable with RSP3 as best enhancer and antagonist followed by RSP5 > RSP7 > RSP8. Siderophore mediated antagonism when iron ≤ 20 µM and maximum shoot and root length and dry weight were observed with Pseudomonas as inoculants suggesting application of siderophore producing plant growth promoting rhizobacterial strains in crop productivity.
KEYWORDS: Antagonism Biocontrol; ; Fusarium wilt; Siderophore PGPR;
Download this article as:| Copy the following to cite this article: Sah S, Sahgal M, Singh R. Concomitant Ability of Siderophore Against iron Paucity and Fusarium wilt in Lycopersicon Esculentum. Biosci Biotech Res Asia 2017;14(1). |
| Copy the following to cite this URL: Sah S, Sahgal M, Singh R. Concomitant Ability of Siderophore Against iron Paucity and Fusarium wilt in Lycopersicon Esculentum. Biosci Biotech Res Asia 2017;14(1). Available from: https://www.biotech-asia.org/?p=21552 |
Introduction
Iron is one of the most abundant and major component for various vital functions (photosynthesis, enzyme cofactor, redox reagent, respiration, synthesis of nucleosides and amino acids) of the plant. It has different redox activities and ability to form co-ordination compounds with variety of ligands, forming insoluble hydroxides at neutral and alkaline pH, thus making it unavailable for plants.1 Microbes inhabiting the rhizosphere serve as an intermediary between the plant, which requires soluble inorganic nutrients, and the soil, which contains the necessary nutrients in complex and inaccessible forms.2 In response to evade the solubility problem microbes synthesize iron chelators called siderophores.3
Siderophores are relatively low molecular weight, iron chelating compounds, produced under iron limited conditions in order to enhance the plant growth by scavenging iron from the environment and making the mineral available to the cell near the root.4,5 Siderophore is also known to be one of the biocontrol mechanisms belonging to Plant Growth Promoting Rhizobacteria (PGPR) groups of microbes including Pseudomonas, Azospirillum, Azotobacter, Bacillus, Klebsilla, Entrobacter and Serratia6 by producing antibiotics, antagonistic substances, plant hormones, biocontrol agents7 and bioremediation agents. 8;9;10
Among the PGPR’s fluorescent Pseudomonas spp. has received much attention because of their involvement in natural disease suppressiveness against various bacteria (Ralstonia solanacearum) and phytopathogenic fungi such as Phytophthora parasitica, Phythium ultimum, Fusarium oxysporum veridianthi and Sclerotinia sclerotiorum11;12 suppressing different plant diseases of wheat, black rot of tobacco and Fusarium wilt, fungal diseases of orange, lemon, citrus roots and ornamental plants.13;14 All these studies indicate the effectiveness of fluorescent pseudomonads to control the plant pathogens in different crops. Since F. oxysporum is a common pathogen and responsible for wilting of banana, strawberry, muskmelon, asparagus, spinach, lettuce, tomato plants leading to death,15 to overcome the wilting disease in Lycopersicon esculentum, radish (Raphanus sativus L.), Pythium damping-off and root rot of cucumber and Botrytis cinerea in tomato, Pseudomonas sp. can be used as an ecofriendly biocontrol agent.16
The purpose of the study was to isolate siderophore producing Pseudomonas spp from forest/ agricultural soil, and utilize them for enriching the iron deficient soil. The siderophore producing property of Pseudomonas isolates were further explored for controlling the growth of Fusarum oxysporium. The research could be beneficial in utilizing the properties of Pseudomonas as an effective strategy to enhance iron availability as well as biological control agent.
Material and Method
Field Site
Soil sample were collected from two ecosystems of Uttarakhand State, India; Pinus and Querecus forest of Lamgarah 2,000m and cultivated fields of Almora situated at the southern edge of the Kumaun Hills of the Himalaya range.17
Isolation of Bacteria
Six different soil samples were collected from three sites (Pine forest, Oak forest and Agricultural land) at a depth of 1-10cm surface soil and 1ft deep soil. Soil samples were serially diluted and aliquots of the resulting solutions were plated on King’s B 18 and Gould’s media.19 Plates were incubated at 28oC for 24 hrs.
Screening for Siderophore Producing Isolates
A total of 172 isolated colonies were screened for siderophore production. Colonies from King’s B and Gould’s media plate were inoculated in King’s B broth and incubated for 24 hours at 28oC and 120 rpm. Loop full of overnight grown culture was spot inoculated on Chrome azurol ‘S’ agar media (CAS media) containing: (A) Chrome azurol ‘S’ (B) 1mM FeCl3.6H2O in HCl, (C) HTMA in nutrient agar (pH 7).20 These plates were kept in incubator at 28oC for 48 hrs. Total 51 isolates were found positive on CAS media plate for siderophore production.
Quantification for Siderophore Production
14 siderophore producing isolates with zone size more than 2cm on CAS plate were selected for quantification of siderophore produced. Sodium Succinate Medium (SSM medium) gL-1: K2HPO4– 6.0, KH2PO4 – 3.0, (NH4)2SO4--1.0, MgSO4 .7H2O – 0.2, Succinic acid- 4.0, Agar- 20 (21) was used for quantification with two sets a) SSM supplemented with 20µM Fe, b) SSM without adding 20µM Fe. SSM was inoculated with overnight grown culture of presumptive Pseudomonas and incubated at 28oC for 24hrs at 120 rpm. SSM was then centrifuged at 5000 rpm for 8 min (Sigma 3 K30) and absorbance of the supernatant was read at 400 nm. Samples from SSM medium inoculated with overnight grown culture were withdrawn after 24 hrs. Siderophore produced was quantified using formula.
Siderophore Conc. = Aλ x M.Wt of siderophore / Eλ x 10-6 (mg/L) or (µg/mL)
The data was analyzed for comparative statistics e.g., mean ± standard error, One Way Analysis of Variance (ANOVA), and t-test (two tailed). Values of P < 0.05 were considered as significant. All statistical analyses were performed using Sigma Plot (Systat Software, San Jose, California USA), and IBM SPSS Statistics for Windows, Version 20.0. Armonk, NY: IBM Corp.)
Screening of Pseudomonas isolates for Antagonistic Property Against Fusarium oxysporum
Plate assay method was used to observe the antagonistic property of 14 best siderophore producing Pseudomonas isolates against F. oxysporum. F. oxysporum disc of 6-mm was placed centrally on PDA plates. Sterilized filter paper strips (2mmx8mm) loaded with Pseudomonas sp. (2 x 108 CFU/gm) were placed at a distance of 3cm on the two opposite sides of the disc and incubated for 4 days at 28oC.
Identification of the isolates
The amplification reaction for the isolates RSP7, RSP5, RSP3, RSP8 was performed in a 50 𝜇L volume by mixing template DNA (2 𝜇L), 1 𝜇L (75 pmol/𝜇L) 16S primer forward (5’AGAGTTTGATCATGGCTCAG3’) and reverse (5’TACGGCTACCTTGTTACGACTT3’), 25 𝜇L mastermix (1X, G-Biosciences) containing Taq polymerase, PCR reaction buffer and dNTPs. DNA amplification was done in a DNA thermal cycler (Master cycler pro, Eppendorff) with the temperature profile: initial denaturation at 94oC for 5 min, 40 cycles of denaturation at 94oCfor 30 sec, annealing at 50oC for 30 sec, and extension at 72oC for 1 min, with a final extension at 72oC for 10 min. The amplified product along with DNA molecular weight markers was run on a 0.8% agarose gel mixed with ethidium bromide at a constant voltage (60 v) and visualized in gel documentation system (InGenius3, Synegene). Amplified DNA product was eluted from agarose gel using Qiagen gel elution kit as per the manufacturer’s instructions and protocol. The pure eluted amplified DNA product was sent for 16S rDNA sequencing.
Phylogenetic Analysis and Strain Identification
The obtained 16S rDNA sequence was subjected to nucleotide blast (blastn) at NCBI to retrieve homologous sequences and identify the strain to the generic level. The multiple sequences were aligned using CLUSTALW2, the multiple sequence alignment program from EMBL-EBI, UK, and the phylogenetic tree was constructed through neighbor-joining method in Phylip and viewed using TreeView program.
Antagonistic Potential Against fusarium wilt in Lycopersicon Esculentum (Pot Trial Experiment)
Four Pseudomonas isolates namely RSP7, RSP5, RSP3, RSP8 showing the maximum antagonistic activity and identified as various strains of Pseudomonas aeruginosa were selected for pot trial experiment. Soil and sand were passed through a sieve mesh and autoclaved three times with a day’s gap in between, autoclaved soil and sand were mixed in a ratio of 3: 1 and filled in pots with a size of 100kg each. Disease free Tomato plants were taken from a 15day nursery and the experiments were set up in triplicate. Different combinations were designed in triplicates as: a) Infected soil with fungus; b) Infected soil with bacteria; c) Infected soil with bacteria and fungus; d) Infected soil with bacteria and fungus with Fe (20 µM); e) Inoculation of bacteria to diseased plant f) Control: Without fungus and bacteria.
For inducing disease to the plants F. oxysporum spores (1×107spore ml-1) were mixed with the soil. Plant roots were dipped in the mother culture of selected isolates according to the experimental design and planted. The influence of iron was evaluated by employing Hoagland solution.22 For this, Hoagland solution without 20µM iron was used to induce siderophore production and Hoagland solution with 20µM iron was used to stop/reduce siderophore production. Hoagland’s solution was added twice a week along with sterile distilled water at regular intervals (i.e., alternate days), throughout the duration of the experiment. The experiment was studied under greenhouse, with a temperature of 18oC (night) and 24oC (day), 80% relative humidity for 30 days. After 30 days of plantation shoot length, root length, fresh wt., and dry wt. for all the plants (triplicates) were recorded.
Result and Discussion
Isolation of Bacteria
Fluorescent as well as non-fluorescent Pseudomonas colonies were able to grow in King’s media, as it is relatively nonselective media23 and fluorescent Pseudomonas were able to grow in Gould’s media selectively as the media is based on a detergent, sodium lauryl sarcosine, and an antibiotic, trimethoprim19 thus the number of colonies in both the media differed. A total CFU of 17.2 x 105 were found from all the six sample of soil. Pseudomonas population was found to be dominant as they have a strong competitive behaviour, colonization potential and sustainability in soil/ rhizosphere exerting growth promoting influence on a variety of plant species.24
Screening for Siderophore Producing Isolates
Among 172 colonies on King’s B and Gould’s media, 36 were found to from an orange color halo. Fe (III) gives the agar a rich blue color and concentration of siderophores excreted by iron starved organisms results in a color change to orange. The change in the color of medium from blue to orange halo, confirmed siderophore production on CAS medium25 also effectively differentiated between bacteria that were able to excrete large amounts of siderophore by performing this same method (CAS agar).
Quantification of Siderophore Production with and without adding iron (Fe3+)
Siderophore quantification for 14 siderophore producing isolates was done with and without adding 20 µM iron. Supernatant of SSM medium after 24hrs of inoculation was scanned spectrophotometrically and clear peak in the range of 367- 400nm confirmed the production of siderophore.21 Siderophore production without addition of iron was found to be 74.7-210µg mL-1, while it was observed that after iron supplementation, siderophore production was repressed by 24-86% (Table I). RSP5 showed maximum siderophore production (134.18 µg mL-1 and 210.00 µg mL-1) while RSP8 showed minimum (10.31 µg mL-1and 74.7 µg mL-1) with and without iron respectively. QD2, PS10, RSP3 produced 127.63 µg mL-1, 135.81 µg mL-1 and 197.00 µg mL-1 siderophore without adding iron and siderophore production was reduced to 127.183 µg mL-1 in QD2, 124.81µg mL-1 in PS10, and 135.36 µg mL-1 in RSP3 after adding iron. Siderophore production by RSP7, RSP5 and PS5 without adding iron was almost same as 142.03µgmL-1, 142.08µg mL-1 and 141.72 µg mL-1 respectively but after addition of Fe, 16%, 47% and 24% reduction in siderophore production was observed in RSP7 (119.10 µg mL-1), RSP5 (74.7 µg mL-1) and PS5 (108.7 µg mL-1) respectively.
Table 1: Quantification of siderophore production before and after adding Fe (20µM)
| S.No. | Presumptive Pseudomonas | Quantity of siderophore produced with 20µM Fe (400nm)
|
Quantity of siderophore produced without Fe (400nm)
|
Paired t test value (t)
|
Level of significance (P) |
| 1 | AS17 | 114.18 | 158.63 | 42.327
|
P < 0.001* * * |
| 2 | PS5 | 108.7 | 141.72 | 19.036
|
P < 0.001* * * |
| 3 | QD2 | 127.183 | 127.63 | 0.178
|
P > 0.05 |
| 4 | QD3 | 124.81 | 137.90 | 31.231
|
P < 0.01* |
| 5 | RSP5 | 134.18 | 210.00 | 100.259
|
P < 0.001* * * |
| 6 | RSP3 | 119.36 | 165.00 | 51.197
|
P < 0.001* * * |
| 7 | PDG2 | 105.909 | 133.00 | 12.756
|
P < 0.01* |
| 8 | RSP2 | 135.36 | 197.45 | 65.445
|
P < 0.001* * * |
| 9 | RSPG2 | 106.45 | 199.18 | 62.359
|
P < 0.001* * * |
| 10 | PD5 | 160.45 | 167.81 | 6.519
|
P > 0.05 |
| 11 | PS10 | 124.81 | 135.67 | 20.745
|
P < 0.01* |
| 12 | RSP7 | 119.10 | 142.03 | 28.402
|
P < 0.001* * * |
| 13 | RSP5 | 74.7 | 142.08 | 94.607
|
P < 0.001* * * |
| 14 | RSP8 | 10.31 | 74.7 | 232.105
|
P < 0.001* * * |
Note: Values are a mean of three replicates
* Significant, *** highly significant
Table 1: The table represents quantification of siderophore with and without adding 20 µM iron. The siderophore range after addition of 20 µM Fe was negligible. Thus after addition of Fe (>20 µM) an inverse relationship between the amount of siderophore synthesized and the iron content of the medium. The results were supported with paired t test analysis and its significance (P).
An inverse relationship between the iron content of the medium and the amount of siderophore synthesized was observed suggesting that siderophore production is inversely proportional to iron concentration and siderophore production is inhibited if the concentration of Fe is ≥ 20 µM21;1; 27 have also indicated the presence of siderophore in the same range.
Statistical Analysis
A paired t-test analysis showed that siderophore production by Pseudomonas strain RSP8 after adding 20µM Fe (mean = 10.31µg mL-1) was significantly lower (t=-232.105; mean diff. of 64.40; P< 0.001) than without addition of Fe (mean = 74.7µg mL-1). Similarly, in RSP5 a significant difference was found after adding Fe, as the siderophore production was lowered (mean difference = 68; t=94.60; P< 0.001). Fe reduced the siderophore production significantly in RSP5 (mean difference = 76; t= 100.25; P< 0.001). RSP2, RSP3, RSPG2 responded almost similarly with a t value = 51.197: t = 65.445; t = 62.359 respectively, and showing a significant difference of P< 0.001. Siderophore produced by AS17 was significantly higher (t = 42.327; mean difference = 44.45; P< 0.001) in soils without Fe as supplement (158.63 µgmL-1) than the soils with Fe as supplement (114.18 µgmL-1). Similar trends were also observed in PS5 strain where also the siderophore production proved to be significantly greater (t = 19.036; mean difference = 33.02; P< 0.001) in soils without Fe as supplement (141.72µgmL-1) than the soils with Fe as supplement (108.7µgmL-1). However, QD2 induced siderophore production did not show any significant difference (t = 0.178; mean difference = 0.447; P> 0.05) in soils without Fe as supplement (127.63µgmL-1) than the soils with Fe as supplement (127.183µgmL-1). Similarly, PD5 did not show any significant difference in siderophore production after adding Fe (t= 6.519; mean difference = 7.36; P > 0.05). The statistical analysis also supports that siderophore production is related to presence of iron. A considerable difference was observed for some isolates (RSP8: 86%, RSP5: 47%, RSPG2: 46%, RSP2: 36%, RSP3: 31%, PS5: 23%, AS17: 27%) in presence and absence of Fe, confirming sensitivity of siderophore for iron. The results were found to be in accordance with earlier reports as by.21;27
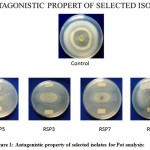 |
Figure 1: Antagonistic property of selected isolates for Pot analysis:
|
Figure 1: Zone of inhibition showed by Pseudomonas isolates RSP5, RSP3, RSP7, RSP8 against Fusarium oxysporium Control Fusarium oxysporium isolate having a zone size of 3.4 cm Different strains of Pseudomonas isolates showing inhibition in the growth of Fusarium oxysporium RSP5 : 48.5% , RSP3 : 37.1 %, RSP7 : 20 %, RSP8 : 0%
Screening of Pseudomonas isolates for Antagonistic Property Against Fusarium oxysporum
The 14 isolates were quantified with and without Fe were screened for their antagonistic property against F. oxysporium by plate assay method (Fig 2). Isolate RSP8 was unable to show antagonism against F. oxysporium as plate was fully covered with F. oxysporium mycelium representing 0% zone of inhibition. Isolate QD2 and PDG2 showed the maximum antagonistic property of 57% while RSP5, RSPG2, AS17 showed 48% antagonism (Table II).
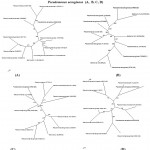 |
Figure 2: Phylogenetic tree based on 16S rRNA gene sequences of four isolated strains of Pseudomonas aeruginosa (A, B, C, D)
|
Figure 2: A neighbor-joining analysis and bootstrap support was performed on the gene sequences. Bootstrap values are given at nodes. Values in parentheses are accession numbers of the isolated strains (KR051489, KR051487, KR051490, and KR051488 respectively).
It was observed that siderophore produced by RSP8 (74.7 µg mL-1) was minimum and was not able to show any antagonism against F. oxysporium (0%). RSP5 produced large amount of siderophore (210.00 µg mL-1) and suppressed the growth of F. oxysporium by 48%. RSP3 produced 197 µg mL-1 siderophore and was able to inhibit 57% growth of F. oxysporium. Isolates RSP7, PS10 were antagonist to F. oxysporium by 20% and 28.5% and had a siderophore production of 142.03, 135.67 µg mL-1 respectively. RSP5 and PS5 having similar amount of siderophore production (142.08, 141.72 µg mL-1 respectively) could suppress the disease by 40%. These results suggest that siderophores apart from their role in active transport of iron also restrict the growth of pathogens.27
Statistical analysis
A paired t test was performed for the results on plate. The t-test values represent the comparison of antagonistic effect shown by isolates to the control. The highest level of antagonism (57%) was shown by QD2 and PDG2 on plate and a t- value of 25 and 7 respectively with a significance of P < 0.05. AS17, RSP5 and RSPG2 represented the second highest antagonism of 48.5% on plate, and the paired t test values a st = 8.4, 14, 3 respectively with a significance of P< 0.01 for AS17 and RSP5 and P< 0.05 for RSPG2. PS5, PD5, RSP5 showed an antagonism of 40% with t values of 62, 3, 10 respectively and significance of P< 0.001, P< 0.05, P< 0.01. RSP7 shows an antagonism of 20% on plate with t value of 31.1 and P< 0.01. Paired t-test analysis proved a highly significant antagonism with isolates RSP7, PS10, QD2, PS5 (t = 37.37, t =18.5; t = 25.90 t = 62 respectively P < 0.001). RSP2 (t=8.2) and RSP3 (t=7.9) represent almost similar t values with a significant antagonism of P < 0.01. RSP3 showed an antagonism of 57.1%. (Table II). The analysis supports our study that the Pseudomonas isolates producing significant amount of siderophore in deficiency of iron possess antagonistic property.
Table 2: Antagonism against Fusarium oxysporium plate assay
| S.No. | Presumptive Pseudomonas | Antagonism against Fusarium oxysporium (cfg) (cm) ±S.E |
% inhibition |
t- value |
Level of significance (P) |
| 1 | AS17 | 1.7± 0.208 | 48.5 | -8.64692 | P<0.01 |
| 2 | PS5 | 1.4± 0.033 | 40 | -62 | P< 0.001 |
| 3 | QD2 | 2.0 ± 0.057 | 57.1 | -25.9808 | P<0.001 |
| 4 | QD3 | 1.5 ± 0.202 | 42.8 | -9.53514 | P< 0.01 |
| 5 | RSP2 | 1.6 ± 0.230 | 45.7 | -8.22724 | P<0.01 |
| 6 | RSP5 | 1.7 ± 0.2 | 48.5 | -14.4222 | P<0.01 |
| 7 | PDG2 | 2.0 ± 0.185 | 57.1 | -7.90263 | P<0.01 |
| 8 | RSP3 | 1.3 ± 0.15 | 37.1 | -7.20056 | P< 0.01 |
| 9 | RSPG2 | 1.7 ± 0.8 | 48.5 | -3.74304 | P<0.05 |
| 10 | PD5 | 1.7 ±0.497 | 40.0 | -3.48218 | P< 0.05 |
| 11 | PS10 | 1.0 ±0.133 | 28.5 | -18.5 | P≤ 0.001 |
| 12 | RSP5 | 1.4 ±0.2 | 40.0 | -10.5 | P< 0.01 |
| 13 | RSP7 | 0.7 ± 0.088 | 20 | -31.3711 | P< 0.001 |
| 14 | RSP8 | 0 ±0 | 0 |
C- Control (3.5) -fg = fungal growth; % inhibition =control-fungal/control *100
Data represent average of three replicates.
Table 2: The table represents the antagonism property of 14 isolates against Fusarium oxysporum by plate assay method. Isolate RSP8 was unable to show antagonism against F. oxysporum (0% zone of inhibition, 10.31 µg /mL siderophore production), isolate QD2 and PDG2 showed the maximum antagonistic property of 57% and RSP5, RSP2, AS17 showed 48% antagonism suggesting that siderophores apart from their role in active transport of iron act as growth antagonists by means of sequestering iron from the environment, restricting the growth of pathogens. The paired t-test support the results and observations with their respective P values (Level of Significance) values.
Pseudomonas sp. inhibits the mycelial growth of many species of Aspergillus, Penicillium and Fusarium.28 The biocontrol properties of the bacteria belonging to the genus Pseudomonas are considered better because of their adaptive metabolism and their ability to produce an array of compounds inhibiting the growth of several fungal pathogens. Pseudomonas sp. has been reported for its dominance in soils of colder climatic conditions of the Himalayas and its role in plant growth promotion and biocontrol.29 Several Pseudomonas species have been extensively used for biological control against many soil-borne plant pathogens.11
Identification of the isolates
The blast studies performed with sequence of the amplified 16s rDNA of the four isolates RSP7, RSP8, RSP3 and RSP5 after purification and sequencing showed that the strain exhibited 93.0–99.0% similarity with different Pseudomonas species and 99% similarity with various strains of Pseudomonas aeruginosa. Thus on the basis of molecular studies the Pseudomonas strains were identified as a Pseudomonas aeruginosa. The GenBank /NCBI accession number of the strain Pseudomonas aeruginosa strains were as – RSP3 – KR051489 (Fig 3A), RSP7 – KR051487 (Fig 3B), RSP8 – KR051488 (Fig 3C) and RSP5 – KR051490 (Fig 3D)
 |
Figure 3: Phylogenetic tree based on 16S rRNA gene sequences of Pseudomonas aeruginosa along with the Pseudomonas aeruginosa and other closely related Pseudomonas species:
|
Figure 3: A neighbor-joining analysis and bootstrap support was performed based on 16S rRNA gene sequences of Pseudomonas aeruginosa along with the Pseudomonas aeruginosa and other closely related Pseudomonas species mainly with the seven subclusters of Pseudomonas sensustricto group, P. syringae group, P. chlororaphis group, P. fluorescens group, P. putida group, P. stutzeri group, P.aeruginosa group and P. pertucinogena group. Bootstrap values are given at nodes. Values in parentheses are accession numbers (KR051487, KR051488, KR051489 and KR051490)
Phylogenetic Analysis
The phylogenetic tree showed the detailed evolutionary relationships between the newly identified strain Pseudomonas aeruginosa and other closely related Pseudomonas species mainly with the seven subclusters of Pseudomonas sensu stricto group30 P. syringae group, P. chlororaphis group, P. fluorescens group, P. putida group, P. stutzeri group, P. aeruginosa group and P. pertucinogena group. It was observed that all of these four isolates were in close association with each other. Among the seven groups all the isolates were similar to P. aeruginosa group and demonstrated a distinct phylogenetic position of this strain within the genus (Fig4).
 |
Figure 4: Results of the pot analysis experiment
|
Figure 4: Assessment of siderophore mediated antagonistic effect against Fusarium wilt in L. esculentum FUS – Fusarium oxysporium; Fe – Iron; Bacterial (Pseudomonas) isolates: RSP3, RSP5, RSP7, RSP8
Antagonistic Potential Against fusarium wilt in Lycopersicon Esculentum (Pot Analysis)
Based on the results of plate assay method and quantification of siderophore produced in presence and absence of iron, four Pseudomonas aeruginosa strains (RSP7, RSP8, RSP3 and RSP5) were selected for assessment of siderophore mediated antagonistic effect against Fusarium wilt in L. esculentum (Fig 5). After 30 days of pot trial experiment, it was observed that the treated plants were sturdier and taller than the control plants even at the early stages of growth (Fig 5). When soil was inoculated with RSP3 the shoot length, root length, fresh weight and dry weight of plant was maximum with increase of 44%, 64%, 50%, and 35% respectively. Appreciable difference of 82% increase in fresh weight was observed when the plant was inoculated with F. oxysporum along with RSP3. Iron along with RSP3 and F. oxysporum decreased siderophore production but still the root length of plant increased by 50%. RSP8 neither proved to be an effective enhancer for plant growth nor as a good antagonist with a decrease in root length, fresh weight and dry weight with mere increase of 18% in dry weight. The fresh weight further decreased to 55% in presence of iron. The results among the four isolates are comparable with RSP3 as best enhancer and antagonist followed by RSP5> RSP7>RSP8 (Table III). Earlier (Rao et al., 1999 (36) also studied the influence of five strains of fluorescent Pseudomonads (GRP3, GRP6, PRS9, RBP2 and PEn4) on growth and nodulation of lentil in a Fusarium infested soil and found that the strains PRS9 and GRP3 were found to reduce the population level of pathogen by 25-50 and 50-75 per cent in rhizosphere and rhizoplane respectively.
Table 3: Plant growth characteristics after 30 days pot trial experiment
| Experimental strains used | Shoot Length (cm) | Root Length (cm) | Fresh wt.
(gm) |
Dry wt.
(gm) |
| RSP5 | 10.50 ±1.323 | 5.33 ±0.333 | 2.99 ±1.50 | 0.38 ±0.081 |
| RSP3 | 10.83 ±0.928 | 4.66 ±0.289 | 2.33 ±2.30 | 0.27 ±0.042 |
| RSP7 | 8.33 ±0.882 | 5.50 ±0.289 | 1.46 ±1.47 | 0.19 ±0.028 |
| RSP8 | 8.00 ±0.764 | 2.66 ±0.289 | 2.16 ±2.17 | 0.18 ±0.044 |
| RSP5+FUS | 8.33 ±1.093 | 2.50 ±0.500 | 1.99 ±1.90 | 0.25 ±0.056 |
| RSP3+FUS | 10.83 ±1.641 | 3.50 ±0.289 | 1.88 ±1.87 | 0.14 ±0.021 |
| RSP7+FUS | 7.40 ±0.208 | 2.16 ±0.167 | 2.22 ±2.20 | 0.13 ±0.027 |
| RSP8+FUS | 6.33 ±0.333 | 2.00 ±0.577 | 0.87 ±0.87 | 0.07 ±0.029 |
| RSP5+FUS+Fe | 2.50 ±0.500 | 2.00 ±0.289 | 0.23 ±0.23 | 0.13 ±0.012 |
| RSP3+FUS +Fe | 5.33 ±0.167 | 3.00 ±0.000 | 0.84 ±0.85 | 0.28 ±0.114 |
| RSP7+FUS +Fe | 4.50 ±0.289 | 2.66 ±0.333 | 1.68 ±1.68 | 0.14 ±0.042 |
| RSP8+FUS+Fe | 2.00 ± 0.577 | 1.55 ±0.000 | 0.48 ±0.49 | 0.13 ±0.111 |
| FUS | 4.16 ±0.167 | 2.00 ±0.000 | 1.03 ±1.03 | 0.11 ±0.003 |
| CONTROL | 7.50 ±0.167 | 2.83 ±0.441 | 1.55 ±2.90 | 0.20 ±0.035 |
Note: Values are a mean of three replicates
FUS- Fusarium oxysporium
Fe – Iron
Table 3: The table represents the results of the pot trial experiment in presence and absence of 20 µM iron. The results among the four isolates are comparable with RSP3 as best enhancer and antagonist followed by RSP5 > RSP7 > RSP8.The results were based on the shoot length, root length, fresh weight and dry weight of plants.
It was also observed that siderophore production is beneficial in combating the disease produced by F. oxysporium as strains producing good amount of siderophore are effective in protecting the plant of Lycopersicon esculentum from the Fusarium wilt. For example, siderophore produced by RSP3 was found to be highest (197.45µgmL-1), and the plants grown in presence of F. oxysporium along with RSP3 were sturdier while siderophore produced by RSP8 was minimum (74.7 µgmL-1) and was not able to overcome the disease. Siderophore produced by RSP3 was utilized by the plant to combat the disease, suggesting that the siderophore produced by Pseudomonas helps the plants to overcome the Fusarium wilt of L. esculentum. Properties of Pseudomonas isolate RSP3 can be effectively used for combating Fusarium wilt and also for enhancing the growth of plants. (Gull and Hafeez, 2012 (37) also reported that siderophore production is the key mechanism involved in the antagonism against pathogenic fungus.
Conclusion
Pseudomonas sp. is ubiquitous bacteria in soil and has many traits along with increasing the availability of iron from soil with the help of siderophore, making them well suited as PGPR. Depending upon the microbial count, the availability of nutrient to plant also changes as microorganisms convert the nutrient to readily available form for the plant. A dominance of Pseudomonas was observed in the soil of Oak and Pine forest, and agriculture soil. Siderophore producing ability of Pseudomonas is sensitive to the iron content. In scarcity of iron, siderophore production increases and as the iron content was ≥ 20µM siderophore production decreases. Siderophore produced by Pseudomonas aeruginosa were effective in combating wilting of L. esculentum. Antagonism increases with siderophore production indicating the inhibitory property of siderophores and can reduce crop yield losses caused by fungi in the root environment. Though different mechanisms might be responsible for the inhibition of F. oxysporium but through our study we found that the siderophore production is one of the main bio-control mechanism associated with the antagonistic potentiality of the rhizobacterial isolates from Kumaun Hills of Uttarakhand.
References
- Bholay A. D., Priyanka J. U., Borkhataria B. V., Mayuri V. D. Fluorescent Pseudomonads as Plant Growth Promoting Rhizobacteria and Their Siderophoregenesis. J. of Pharmacy and Biol. Sc. 2012;3:27-32.
- Pandey A., Palni L. M. S. The rhizosphere effect in trees of the Indian central Himalaya with special reference to altitude. App Eco Env Res. 2007;93:102.
CrossRef - Neilands J. B. Siderophores Structure and Function of Microbial Iron Transport Compounds. J. Biol. Chem. 1995; 270(45):26723-26.
CrossRef - Hider R. C and Kong X. Review Chemistry and biology of siderophores. Natural Product Reports. The Royal Society of Chemistry. 2010;27:637–657.
- Ahmed E., Holmstrom S. J. Siderophores in environmental research: roles and applications. Microb. Biotechnol. 2014;7(3):196-208.
- Weller D. M., Raaijmakers J. M., Gardner B. B. M., Thomashow L. S. Microbial populations responsible for specific soil suppressiveness to plant pathogens. Annual Review of Phytopathology. 2002;40:308–348.
CrossRef - Verma V.C., Singh S. K., Prakash S. Bio-control and plant growth promotion potential of siderophore producing endophytic Streptomyces from Azadirachta indica A. Juss. Basic Microb. 2011;51:550–556.
CrossRef - Buss H. L., Luttge A and Brantley S. L. Etch pit formation on iron silicate surfaces during siderophore-promoted dissolution. Chem. Geol. 2007;240:326–342.
CrossRef - Shirvani M., Nourbakhsh F. Desferrioxamine-B adsorption to and iron dissolution from palygorskite and sepiolite. Appl. Clay Sci. 2010;48:393-96.
CrossRef - Ishimaru Y., Takahashi R., Bashir K., Shimo H., Senoura T., Sugimoto K., Ono K.,Yano M., Ishikawa S., Arao T., Nakanishi H., Nishiza W. A. Characterizing the role of rice in Manganese, Iron and Cadmium Transport. Sci Rep. 2012;2:286-88.
CrossRef - Whipps J. M. Microbial interactions and biocontrol in the rhizosphere. J. Exp. Bot. 2001;52:487-511.
CrossRef - Wahyudi A. T., Astuti R. P., Widyawati A., Meryandini A., Nawangsih A. A. Characterization of Bacillus sp. strains isolated from rhizosphere of soybean plants for their use as potential plant growth for promoting Rhizobacteria. J. Micro. Antimicrob. 2011;3(2):34-40.
- Thomashow L. S., Weller D. M., Bonsall R. F., Pierson L. S. Production of the antibiotic phenazine-1-carboxylic acid by Fuorescent Pseudomonas species in the rhizosphere of wheat. Appl. Environ. Microbiol. 1990;56:908-912.
- Raaijmakers J. M., Leeman M., Oorschot M. M. P. V.,der Sluis I. V., Schippers B., Bakker P. Dose-response relationships in biological control of Fusarium wilt of radish by Pseudomonas spp. Phytopathol. 1995;85:1075-1081.
CrossRef - Snyder W. C., Hans H. N. Fusarium oxysporum f. sp. lycopersici (Sacc.) Prepared by Mui-Yun Wong. Soilborne Plant Pathogen Class Project, Spring. 2003;728
- Buysens S., Heungens K., Poppe J., Hofte M. Involvement of pyochelin and pyoverdine in suppression of Pythium induced damping-off of tomato by Pseudomonas aeruginosa 7NSK2. Appl Env. Microbiol. 1996;62:865–87.
- Sah S., Singh R. Effect of Pseudomonas on micronutrient status of forest and agricultural soil of Uttarakhand. Eco. Env. Cons. 2016;22:279-S284.
- King E. O., Ward M. K.., Raney D. E. Two simple media for the demonstration of payociamin and fluorescein. J. Lab. Clin. Med. 1954;44:301-307.
- Gould W. D., Hagedorn C., Bardinelli T. R., Zablotowicz. New selective media for enumeration and recovery of fluorescent pseudomonads from various habitats. Appl Env. Microbiol. 1985;49:28-32.
- Schwyn B., Neilands J. B. Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 1987;160:47-56.
CrossRef - Meyer J. M., Abdallah M. A. The fluorescent pigment of Pseudomonas fluorescens: Biosynthesis purification and physiochemical properties. J. Gen. Microbiol. 1978;107:319–328.
CrossRef - Hoagland D. R., Arnon D. I. The water-culture method for growing plants without soil. California Department of Agriculture experimental Station Circular. 1950;347.
- Sands D. C., Rovira A. D. Isolation of fluorescent pseudomonads with a selective medium. Applied microbiol. 1970;20(3):513.
- Glick B. R. The enhancement of plant growth by free living bacteria. Can. J. of Microbiol. 1995;41:109-117.
CrossRef - Frey-Klett P., Chavatte M., Clausse M. L., Courrie S., Roux C. L., Raaijmakers J., Martinotti M. G., Pierrat J. C., Garbaye J. Ectomycorrhizal symbiosis affects functional diversity of rhizosphere fluorescent pseudomonads. New phytologist. 2005;165:317-328.
CrossRef - Nair A., Juwarkar A. A.., Singh S. K. Production and characterization of siderophores and its application in arsenic removal from contaminated soil. Water Air and Soil Pollution. 2007;1:199-212.
CrossRef - Berg G., Hurze S., Boucher A., Wellington E. M., Smalla K.. Successful strategy for the selection of new strawberry-associated rhizobacteria antagonistic to Verticillium wilt. Can. J. Microbiol. 2000;46:1128-1137.
- Nourozian J., Etebarian H. R and Khodakaramian G. Biological control of Fusarium graminearum on wheat by antagonistic bacteria, Songklanakarin. J. Sci. Technol. 2006;28:29-38.
- Pandey A., Palni L. M. S., Hebbar K. P. Suppression of damping off in maize seedlings by Pseudomonas corrugata. Microbiol. Res. 2001;156:191–4.
CrossRef - Peix A., Ramírez-Bahena M. H., Velázquez E. Historical evolution and current status of the taxonomy of genus Pseudomonas. Infection Genetics and Evolution. 2009;9(6):1132-47.
CrossRef - Gull M., Hafeez F. Y. Characterization of siderophore producing bacterial strain Pseudomonas fluorescens Mst 8.2 as plant growth promoting and biocontrol agent in wheat. African J. of Microbiol. Res. 2012;6(33):6308-18.
CrossRef

This work is licensed under a Creative Commons Attribution 4.0 International License.





