Manuscript accepted on : 25-06-2021
Published online on: 27-07-2021
Plagiarism Check: Yes
Reviewed by: Dr. Jurdas Sezirahiga
Second Review by: Dr. Monica Butnariu
Final Approval by: Dr. Fernando Lidon
Flavonoid Biosynthetic Pathway: Genetics and Biochemistry
Ramanjeet Kaur, Lubna Aslam, Shajaat Hussain, Nisha Kapoor and Ritu Mahajan*
and Ritu Mahajan*
School of Biotechnology, University of Jammu, Jammu, Jammu and Kashmir, India
Corresponding Author E-mail: ritufeb@gmail.com
DOI : http://dx.doi.org/10.13005/bbra/2914
ABSTRACT: Plants are sessile organisms which are capable of producing a large array of metabolites, required for their adaption and survival. Flavonoids are low molecular weight metabolites with C6–C3–C6 carbon backbones and are categorised into different classes on the basis of structural organization and polymerization. The biosynthesis and distribution of flavonoids depends on the development stage of the plant as well as on diverse environmental conditions. They play a significant role as pigments, phytoalexins, attractants of pollinators and promotes auxin transport. In plants, antioxidant and antimicrobial activities are attributed to interaction of flavonoids with various enzymes, transcription factor and signalling pathways. This review aims to provide the current understanding of structure, their types, biosynthesis and regulation of flavonoid pathway that provide the insights to the key regulating factors and their interactions which makes them the most promising and interesting targets for plant breeding programs to enhance the value-added products in plants. In this review the deep knowledge of flavonoid regulation by micro-RNAs has been provided that attracts the biotechnologists to develop new molecular approaches so as to engineer various plant metabolic pathways to enhance the health-promoting metabolites in plants for human consumption.
KEYWORDS: Flavonoids; Gene Expression; miRNAs; Transcription Factors
Download this article as:| Copy the following to cite this article: Kaur R, Aslam L, Hussain S, Kapoor N, Mahajan R. Flavonoid Biosynthetic Pathway: Genetics and Biochemistry. Biosci Biotech Res Asia 2021;18(2). |
| Copy the following to cite this URL: Kaur R, Aslam L, Hussain S, Kapoor N, Mahajan R. Flavonoid Biosynthetic Pathway: Genetics and Biochemistry. Biosci Biotech Res Asia 2021;18(2). Available from: https://bit.ly/2UVXP70 |
Introduction
Plants and their extracts have been used in traditional medicines for the treatment of various ailments as they have less or no side effects. Plants being sessile produce structurally diverse secondary metabolites so as to adapt themselves to various environmental conditions. Most of these metabolites are the natural end products of the primary metabolism1. They include coumarins, phenolics, tannins, lignins, isoflavonoids and flavonoids. Out of several metabolites, flavonoids are predominant and ubiquitously present in the plant kingdom2. These are the low molecular weight polyphenolic compounds that are not involved in the growth3. The unique physio-chemical properties of flavonoids enable their interaction with different targets, thus influencing various biological functions in plants4,5.Apart, they play a key role in imparting flower colour to various plant tissues.Even, Mendel used flower pigmentation as one of the major characters for the elucidation of inheritance of traits in genetics. Similarly, McClintock observed and reported the transposable elements that modulate the transcription of flavonoid genes, thus leading to discovery of gene silencing mechanism in maize kernels6.
Flavonoids are grouped into various families based on their structure, degree of polymerization and hydroxylation7. These include chalcones, flavonols, flavanones, flavones, isoflavones and anthocyanins8 (Figure1). These compounds are synthesised by the flavonoid branch of the large phenylpropanoid pathway that leads to the formation of various phenylpropanoids including lignins, flavonoids, stilbenes, phenolics and isoflavonoids9. These compounds are formed by sequential elongation and cyclization of phenylalanine which is a product of shikimate pathway10,11. Also, several downstream enzymes such as chalcone isomerase (CHS), flavanone 3-hydroxylase (CHI), dihydroflavonol reductase (DFR), anthocyanidin synthase (ANS) and UDP- glucose: flavonoid glucosyltransferase (UFGT) results in the formation of different classes of flavonoids (Figure 2). These flavonoids play specific functional roles in plants. Since the development of colour in plants is the result of co-pigmentation processwhich is due to interactions among colourless flavonoids (flavanone and flavonols) with coloured compounds (anthocyanins) but can beincreased by inducing substrate competition between flavonoid genes12. The biosynthesis of flavonoid compounds is controlled by various regulatory proteins that effect their activities and expression. Hence, the biosynthesis of flavonoids is studied in many plants in order tounderstand their role in plant physiology13.
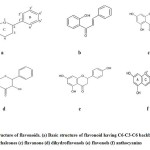 |
Figure 1: Structure of flavonoids. (a) Basic structure of flavonoid having C6-C3-C6 backbone (b) chalcones (c) flavanone (d) dihydroflavonols (e) flavonols (f) anthocyanins |
Various flavonoid biosynthetic enzymes function as multi- enzyme complex that are attached to the endoplasmic reticulum14. In contrast, the accumulation of different flavonoid pigments occurs in either cell wall or vacuole. It was observed that anthocyanins accumulate mostly in the vacuoles which requires a transporter (multidrug resistance associated protein; MRP type)15. The transcription of transporter is co-regulated with the expression of flavonoid structural genes, thus influencing the accumulation of anthocyanins16. In additions, accumulation and transport of flavonoids is also influenced by other factors such as light and circadian clock17.
Flavonoids have key role in plant physiology where it plays major role as pollinator attractors and dispensers, protection against UV radiations, prevent pathogen attack18 and promotes auxin transport19. Every plant has diverse flavonoids profile that makes them unique among species20. This involves biosynthesis of various types of anthocyanins which provide unique flavour and colour to each plant part. They also have nutraceuticals properties that contribute towards human health. Flavonoids compounds have antioxidants as well as pharmacological properties that are responsible for various biological activities in plants21.
Since the flavonoid biosynthesis in plants is the crucial pathway that draws much scientific attention due to contribution to visual appeal to various plant parts and their nutritive value. The complicated metabolic network needs to be deciphered for better understanding the nature of interaction and its role in biosynthesis of different flavonoids. This review covers the major integrated findings related to flavonoid biosynthetic pathway that was studied in last decades. The numerous scientific articles already published in NCBI in last two decades were thoroughly studied and analysed to elucidate the role of key flavonoid genes in plant metabolism. The published reports provide core understanding of the structure-activity relationship of flavonoids and their complex regulation at transcriptional and post-transcriptional level.
Classification and chemistry of flavonoids
Flavonoids are the polyphenolic metabolites that have a benzo-γ-pyrone structure. These are 15-carbon compounds which are derived from a C6-C3-C6 backbone. They have two benzene rings (designated as A and B rings) which are linked by a heterocyclic pyrane ring (designated as C ring) except in chalcones where A and B rings are linearly linked by three carbon chain22. Flavonoids are planar in structure but hydroxylation, methoxylation and glycosylation of hydroxyl group leads to their modification. The degree of hydroxylation at special sites of C6-C3-C6 backbone is favourable for many biological activities of flavonoids23. Flavonoids are classified into different categories depending upon oxidation and degree of unsaturation of the heterocyclic C- ring. These include chalcones, flavones, flavonols, flavanones, dihydroflavonols and anthocyanins.
Chalcones
These are the open chain flavonoids which are common in edible plants. They have A and B aromatic rings that are joined by carbon chain of α, β-unsaturated carbonyl systems which are linear in shape. The formation of chalcones occurs by the condensation of phenylalanine and p-coumaryl CoA24. There are two types of chalcones which are classified on the basis of the presence or absence of OH group at 6th position. The first class is hydroxychalcones (naringenin chalcones) which acts as precursors for the synthesis of downstream classes of flavonoids. The second class is 6’-deoxychalcones which lacks hydroxyl group at 6th position and leads to the formation of 5-deoxy flavonoids and are less abundant in plants25. Production of hydroxychalcones is due to the expression of CHS gene while 6’-deoxychalcones requires expression of both chalcone synthase and chalcone reductase26. The natural occurring chalcones are present in monomeric form which differs from each other in the substitution pattern. The different substituents present are hydroxyl groups, methoxy group, methyl or prenyl groups.
Flavanones and dihydroflavonols
These are formed by the condensation of six membered rings with either α-pyrone or its dihydro-derivatives. Presence of hydroxyl group on 3rd position and double bond on the C-ring differentiates flavonolsfrom flavanones that results in non-planer skeleton of flavanone. Flavanones are formed by the stereospecific cyclization of chalcones that are catalysed by chalcone isomerase (CHI) enzyme which results in the production of 2S-flavanones,which is common substrate in the biosynthesis of various flavonoids. Aglycones and glycosides are the two naturally occurring flavanones present in plants. Dihydroflavonols are the flavanones with a hydroxyl group present at 3rd position. It is a crucial intermediate which bifurcate the flavonoid biosynthetic pathway into two routes. One route lead to the formation of flavonols which is catalysed by leucoanthocyanidin reductase (LAR) and another route results in anthocyanin production catalysed by UFGT gene12. Naringenin is a colourless flavanone which is reduced to dihydroflavonol by the action of DFR enzyme. It catalyses the formation of dihydroflavanol by saturating the C3-C4 double bond27.
Flavonols
These are the flavonoid compounds that are formed by the attachment of hydroxyl group at 3rd position of flavones28. There are two classes of naturally occurring flavonols namely, aglycone and glycosides. There are about 450 types of aglycones and 900 kinds of glycosides. Most of the flavonols exist as O-glycosides which may differ due to the type of sugar moiety attached to them. Flavonols are formed by the oxidation of dihydroflavonols and action of flavonols synthase (FLS) enzyme. The FLS enzyme competes with DFR enzyme for the substrate (2R, 3R-dihydroflavols) for the formation of flavonols29.
Anthocyanins
These are the abundant class of flavonoids responsible for imparting various colours to different plant parts. More than 600 anthocyanins have been identified and reported in various plants30. Apart from colour development, they act as regulators at different development stages, aids in pollination and UV protectants to the plants. The basis of core structure of anthocyanin is 7-hydroxyflavyllium ion. These are 15 carbon compounds (C6-C3-C6) with one fused aromatic ring A, second ring B at position 2 and benzopyran ring C31. The various sugar molecules attached to 3-OH group are present in ring C. Numerous coloured anthocyanins pigments, ranging from yellow to purple colour are due to different glycosylation pattern. The production of anthocyanins is pH dependent and requires metal co-factors for their accumulation in various tissues. Different anthocyanins responsible for colour development in plants are pelargonidin, cyanidin, delphidin and malvidin32.
Flavonoids in plants are mainly in glycosidic form which is mediated by the action of glycosyl transferase. Glycosylation of flavonoids tends to increase their solubility and stability33. Some common glycosylation sites in different flavonoids are: 7-OH in flavones, 3-OH in flavonolsand 5-OH in anthocyanins. Depending upon the attachment of the sugar moiety, flavonoid glycosides are of two types: O-glycosides and C-glycosides. When sugar moiety is attached to the OH group of flavonoid skeleton, it leads to O-glycosides biosynthesis. In contrast, C-glycosides are formed by the attachment of sugar group to the flavonoid skeleton by the C-C linkage34. Similarly, attachment of rhamnose sugar on the 2-OH group of naringenin results in the bitterness of grapes35. Flavonoid glycosides act as phytoalexins and are involved in anthocyanin and proanthocyanidins formation36.
Flavonoid biosynthetic pathway
The flavonoid biosynthetic pathway helps in understanding the chemo-diversity and their potential role in plant development.The first report on isolation of flavonoid biosynthetic genes was from Arabidopsis thaliana37. Flavonoids are produced by phenylpropanoid pathway which in turn is tightly linked to shikimate acid pathway that forms an important aromatic amino acid, phenylalanine. Flavonoid biosynthesis is controlled and regulated by various structural genes and regulatory genes38.
Flavonoid biosynthetic enzymes function as multi- enzyme complex that are attached to the endoplasmic reticulum, while the accumulation of different flavonoid pigments occurs in either cell wall or in vacuole. Anthocyanins are accumulated mostly in the vacuole as they require a transporter, multidrug resistance associated protein. The transcription of transporter is co-regulated with the structural genes expression thatcontrols theanthocyanin accumulation39. Some other factors like light and circadian clock also influences the accumulation and transport of flavonoids40. Several transcription factors modulate thestructural gene transcription, thus regulating the flavonoids accumulation in various tissues. These regulatory enzymes are MYB, bHLH and WD40 transcription factors that mediate their functions by forming a ternary MYB-bHLH-WD40 (MBW) complex41. In addition, some non-coded RNAs (miRNAs) also post-transcriptionally regulate flavonoid biosynthesis.
Chalcone synthase
Chalcone synthase (CHS) belongs totype III polyketide synthase family that catalyse the first crucialreaction in flavonoid biosynthetic pathway. This enzyme functions as a symmetric dimer (where each monomer is ̴ 42KDa polypeptide) and contains two independent active sites. The structure and catalytic machinery of this enzyme first was revealed from the X-ray crystal structure and the functional studies were performed on Medicagosativa CHS protein42. CHS enzyme has conserved cysteine residue that play the role of nucleophile bymediating the movement of intermediates through CoA molecules. Each CHS monomer have upper and lower structural domains. The upper domains contain pseudo-symmetric motif, which is also present in fatty acid β-ketoacyl synthase43. The structural differences between upper and lower domains creates a larger active site that is buried in the interior cavity and provides the space for the intermediates that are involved in chalcone synthesis44,45. The CHS homodimer contains two active sites with each active site containing the residues from a single monomer except Met 137 which is derived from adjoining monomer. Each active site contains Cys 164, Phe 215, His 303 and Asn 336 residues which are responsible for enzymatic function of CHS46. Cysteine 164 play the role of nucleophile by shutting polyketide intermediate while His 303 functions as a general base catalyst resulting in formation of a nucleophilic thiolate anion. Phe 215 and Asn 336 function during decarboxylation reactions thus providing the Van-der Waals interactions favouring the release of CO2 molecules47.
CHS enzyme catalyses the three-step reaction, which involves p-coumaroyl CoA and malonyl CoA condensation for the production of naringenin chalcones (4,2’,4’,6’–tetrahydroxy chalcone). The first step involves the loading of p-coumaroyl moiety from the CoA to the Cys16448. The second step includes a decarboxylation reactionwhere malonyl moiety (obtained from the malonyl CoA) binds with the carbonyl group of coumaroyl thioester. This creates a non-polar environment and facilitates the removal of CO2. The final step involves an intramolecular Claisen condensation wherethree acetate units condense with coumaroyl moiety. This results in the formation of tetraketide intermediate that further undergoes subsequent breakdown and aromatization to yield chalcones49
Chalcones are the important secondary metabolites that along with anthocyanin biosynthesis are also involved in biosynthesis of antimicrobial phytoalexins and flavonoid inducers. These provide plant defence during microbial attack. CHS promoter sequence was first studied in petunia that helped in understanding gene expression pattern in different tissues50. Later, CHS sequences were characterized in several plants like raspberry51, Arabidopsis thaliana45, crabapple52 and mulberry53. The expression of CHS gene has revealed its pivotal role inflavonoid formation during plant development54-57.
Chalcone isomerase
CHI catalyse the intramolecular cyclization of the chalcones into flavanones58. Presence of 2’OH in chalcone is the perquisite requirement for intramolecular cyclization which is mediated by CHI enzyme. The reaction mechanism begins with the formation of 2’-oxyanion that resulted due to the loss of proton from the 2’OH group of chalcones. The newly formed 2’-oxyanion attacks α, β-unsaturated double bond of the substrate resulting in the formation of 2’acidic OH group59. Mechanistic studies revealed that the deprotonation of the chalcone 2’- hydroxyl group is pH dependent and the formation of flavanone is a diffusion limited reaction. Presence of van-der wall forces and extensive H-bonding in the active sites mediates the formation of flavanones.
CHI was first isolated from pea60and is grouped in two types namely, Type I CHI and Type II CHI61. Type I CHI catalyses the conversion of hydroxychalcone into hydroxyflavanone and is ubiquitously present in plant kingdom. Type II CHI catalyses the conversion of both 6’-hydroxychalcone and 6’-deoxychalcone into 5-hydroxyflavanone and 5-deoxyflavanone respectively. CHI is mostly present in leguminous plants. The size of CHI enzyme ranges from 35-465 amino acid residues having molecular weight 23-26 KD.
CHI has been cloned using conserved primers in order to understand its expression profile in different tissues during fruit development. In Arabidopsis, CHI enzyme plays a role of enhancer in flavonoid biosynthetic pathways thus improving the accumulation of flavonoids62, while in Chinese water-chestnut it functions as a promoter that controls its ripening, thus playing an important role in development of this fruit63.
Flavanone 3-hydroxylase
F3H belonging to dioxygenase family catalyse bifurcation of flavonoid pathway into two branches, anthocyanin and flavonols branch64. It catalyses the stereospecific hydroxylation of flavanones to form different types of dihydroflavonols65. Amino acid analysis revealed that the conserved amino acid motifs (His 233, Asp 235, His 289, Arg 299 and Ser 301) binds to the Fe11 ions and 2-oxogluarate thatmediate the redox reaction. F3H belongs to 2-oxoglutarate dependent dioxygenase family, as revealed by three dimensional studies and is based on its absolute requirement of 2-oxoglutarate, molecular O2, ferrous ion and ascorbate for the complete activity of the enzyme66.
Gene expression of F3H has been studied in several plants. In-vitro enzyme assay studies indicated that F3H is effective in catalysing the conversion of naringenin into dihydro kaempferol, thus proved the participation of F3H in the flavonol biosynthesis pathway. Also, the subcellular localization studies confirmed the presence of F3H in the nucleus and cytosol as these sites leads to stabilization of distorted core of 2-ODD and protein-protein interactions67. F3H being a non-heme iron protein, has a significant role in the post translational processing of collagen, plant hormone biosynthesis (gibberlins) and production of β- lactam antibiotics68,69.
Dihydroflavonol-4-reductase
DFR is an oxidoreductase enzyme that stereo-specific reduce thedihydroflavonols into leucoanthocyanidins (Flavan-3,4-diol) which serve as the substrates for the formation of anthocyanidin and proanthocyanidins11. DFR results in the production of colourless, unstable leucoanthocyanidin (leucopelargonidin, leucocyanidin and leucomyricetin). DFR also affects the biosynthesis of other flavonoid compounds such as flavonols and proanthocyanidins. This enzyme controls the carbon flux during flavonoid biosynthesis.
The activity of DFR was first studied and reported in maize plant70. DFR displays a substrate specificity that leads to the production of three different derivatives of anthocyanin pigments such as delphinidin, cyanidin and pelargonidin. The structural information of DFR gene was confirmed by predicting the crystal structure on the basis of its sequence which confirmed that DFR belongs to the short chain dehydrogenase and reductase superfamily that includes YXXXK motif, a C- terminal domain having 4α helices and N-terminal glycine rich motifs that adopts rosamann fold71. This rosamann fold spans the cleft that acts as coenzyme binding site. DFR display different degree of hydroxylationwhich is due to the substrate specificities provided by specific amino acids72. Amino acid Asp134 make DFR enzyme to accept dihydrokaempferol as a substrate whereas presence of aspartic acid showed increased preference for dihydroquercetin73.
UDP glucose: flavonoid 3-O- glucosyltransferase
In plants, glycosylation plays an important role during the biosynthesis of secondary metabolites. UDP glycosyltransferase (UGT) catalyse the glycosylation of various compounds which determine their availability and activity in plants27. Crystal structure analysis predicted that this enzyme belongs to the family 1- glycosyltransferase superfamily, on the basis of presence of GT-B fold which is a major characteristic of this superfamily74. It has two N and C terminal domains with a central β sheet and α helices on its sides. The two domains form a cleft that acts as a binding site for substrate. The active site has highly conserved histidine and aspartate residues which interact with the acceptor thus resulting in the formation of an acceptor-His-Asp complex,that forms β- glucosidic linkage product75. Flavonoids are mostly in glycosylated forms withdifferent sugar donor specificities. Glycosylation of flavonoid quercetin results in different sugar moiety at different positions thus forming 300 different quercetin glycosides, each with different bioactivity76.
The main form of UGT found in plants is UDP-glucose: flavonoid 3-O-glycosyltransferase (UFGT) which has a major role in anthocyanin formation. UFGT is a decisive enzyme which attach sugar molecules to the C3- OH group of anthocyanidin in a direct displacement SN2- like mechanism that leads to formation of coloured pigments12. Colour diversity of anthocyanin depends on degree of hydroxylation on B ring. The important anthocyanins are cyanidin-3-glucoside, cyanidin-3-O rutinoside and cyanidin-3-O galactoside. UFGT geneis important structural gene whose up-regulation is closely associated with the accumulation of anthocyanin in various plant parts.
UFGTdisplay human health promoting benefits such as antioxidant and anticancerous activities77. MYB gene being a regulatory gene binds to the promoter region of UFGT gene and regulates its activity78. The expression of UFGT geneis also up-regulated by various plant hormones and sugars79. Plant hormones such as abscisic acid and ethylene increases the transcript levels of UFGT gene, resulting in colour variegation80.
Flavonoid biosynthesis regulation by MBW complexes
The flavonoid biosynthesis involves early biosynthetic genes and late biosynthetic genes on the basis of structural gene expression and their regulation by various transcription factors. These structural genes are co-ordinately regulated by a MBW complex which consist of three transcription factors namely, basic helix loop helix, MYB transcription factors and WD40 proteins81. These three transcription factors are highly conserved among plants species and well-studied in various model and non-model plants82. MBW complex predominately regulates the late biosynthetic genes, but the degree of regulation differs from species to species and even within tissues83.
The largest family of transcription factors are Myeloblastosis transcription factors (MYB TFs) which are present in all eukaryotes. MYB TFs regulates flavonoid biosynthesis at transcriptional level. These MYB TFs were first characterized from Avainmyeloblastosis virus that have oncogene v-myb and its cellular homologs c-myb84,85. In plants, Colorless1 (C1) was first MYB TF that was identified in maizewhich regulate anthocyanin pigments formation86,87. MYB TFs has a DNA binding MYB domain that acts as transcriptional regulatorin flavonoid biosynthetic pathway. The specificity of MYB domain is due to presence of four imperfect tandem repeats (R) which classify MYB TF into four classes namely 1R-MYB, R2R3-MYB, 3R-MYB and 4R-MYB88. In plants, only R2R3- MYB family is predominant among all the four classes89. R2R3- MYB TFs have been extensively characterized in Arabidopsisfrom where 126 R2R3-MYB TFs have been reported but only few of them act as regulators for flavonoid biosynthesis90. In addition to Arabidopsis, petunia and snapdragon are used for studying flavonoid biosynthesis in plants.
In petunia, bHLH and MYB genes are encoded by Anthocyanin 1 (AN1) and Anthocyanin 2 (AN2) / AN4 respectively that are involved in the biosynthesis of anthocyanins in the floral tissues91. Another R2R2-MYB factor, DEEP PURPLE and PURPLE HAZE also regulates the pigmentation of floral and vegetative tissues. In Antirrhinum, three MYB genes known as Rosea1, Rosea2 and Venosa regulate the flavonoid biosynthetic pathway92. While in apple, three MYB genes (MdMYB1, MdMYB10 and MDMYBA) are co-expressed with bHLH factors to regulate the flavonoid biosynthesis in peel and flesh93. MYB proteins play diverse role in regulation of plant metabolism, stress responses and regulation of cell cycle94.
In addition to MYB TFs, two other transcriptional factors, bHLH and WDR TFs lead to combinatorial regulation of flavonoid biosynthesis in plants. The first bHLH gene was identified in maize which was encoded by the Red 1 locus and is responsible for anthocyanin formation95. Several bHLH genes alongwith R2R3-MYB TFs coordinatethe transcription of flavonoid biosynthetic genes. The major characteristics of bHLH proteins are DNA binding domain, called bHLH domain that binds to specific DNA sequence. It has N terminal end consisting of 18 hydrophilic and 6 basic amino acid residues. In Arabidopsis, 162 bHLH TFs have been reported which are classified into 12 major groups and various subgroups96. In addition, bHLH proteins also regulate plant growth and development, cell patterning and regulation of various hormones signalling pathways97.
WD repeat motif is the third transcription factor of MBW complex which is also known as WD40 as it contains 40 amino acid structural repeats of tryptophan-aspartic acid at its end. WD40 motif was first identified in G proteins that provide a platform for protein-protein interactions98. WD40 TFs have been reported in plants likeArabidopsis (TTG1), maize (PAC1), apple (MdTTG1) and grapes (VvWDR1/2).
Post-transcriptional regulation based on Micro-RNAs
In the recent years, research has been diverted towards understanding the post transcriptional regulation of the flavonoid pathways. There are two classes of endogenous non coding, small regulatory RNAs namely, small interfering RNAs and micro RNAs 99,100. Among these non-coding RNAs, micro RNAs (miRNAs) are the non codingribo-regulators, 21-24 nucleotides in length that regulates the gene expression in eukaryotes. Several studies have conducted to understand the potential role of small RNAs in various processes of plant growth and development101,102. miRNA biogenesis is a step wise process where miRNA gene undergoes transcription and splicing to form pri-miRNA. After loading into RISC complex, it regulates the expression of its target genes post- transcriptionally. Micro-RNA acts as negative regulator by affecting the translation of coding mRNAs that showed perfect or near complementarity103.
miRNAs regulatevarious metabolic processes, biosynthesis of secondary metabolites, hormone signalling and various stress responses104. More than 21 miRNA families reported are highly conserved in angiosperms such as miR156, miR159 and miR160. miR828 was first time reported in Arabidopsis and later, it has been also studied in many plants such as apple and popular. In Arabidopsis, miR828 regulates the anthocyanin accumulation with respect to phosphate deficiency105. Since, miR828 sequence is complementary to MYB transcription factor, hence it provides evidence for its key roles in regulating anthocyanin formation106. Over-expression of miR828 also down-regulates the expression of various MYB genes resulting in reduced level of flavonoid accumulation in Arabidopsis. In tomato, the small tandem target mimic constructs were generated that blocked the miR858, leading to increase in MYB7 transcript levels107. Similarly, in apple and Arabidopsis, cleavage sites of miR828 and miR858 were observed in R3 domain of MYB transcription factor that target various MYB encoding genes thus suggesting the potential role of miR858 in anthocyanin biosynthesis regulation.
Squamosa promoter binding protein like (SPL) proteins play significant role in various processes of plant development including embryonic development, plant fertility and flowering induction108. These SPL9 proteins regulate the flavonoid biosynthesis by binding to the promoter region of regulatory genes109. An increase in the transcript levels of SPL gene along with the growing stem of Arabidopsis was due to decline in the transcription of miR156 during plant development110. In Arabidopsis, anthocyanin rich tissues contain higher levels of miRNA156 which in turn reduces the SPL9 activity. The reduction of miRNA156 targeting SPL9 activity is inversely correlated with the transcript levels of various flavonoid genes, thus affecting the accumulation of anthocyanins111. Therefore, increased expression of SPL genes results in reduced accumulation of anthocyanins and increased flavonol biosynthesis. Competition of SPL9 with bHLH transcription factor for their binding to PAP1 results in destabilization of the MBW transcriptional complex112. This in turn leads to the down-regulation of DFR and UFGT structural genes, thus leading to reduced anthocyanin production. Thus, an antagonistic relationship was observed between anthocyanin and flavonol formation. The expression of different miRNAshas beenquantified using real time polymerase chain reaction (qRT-PCR) in plant (Table 1). Thus, miRNAs are the potential target genesthat help in investigating their roles in plant growth development and other metabolic processes.
Table 1: miRNAs and their target proteins involved inflavonoid biosynthesis.
| miRNA family | Target protein(s) | Functions | References |
| miR828 | MYB113, C1 | Regulates anthocyanin biosynthesis | 107 |
| miR5538, miR477b | DFR | Regulates activity of flavanone 4-reductase, flower development | 113 |
| miR858 | MYB | Regulates MYB-bHLH-WD40 complex | 114 |
| miR156, miR157, miR159 | SPL | Fruit development and ripening, anthocyanin accumulation | 111, 115 |
| miR171 | ANS | Anthocyanidin biosynthesis | 116 |
| miR535, miR396b,miR166i | FNS | FNS | 116 |
| miR396, miR845, miR829 | CHS | Regulates the formation of naringenin | 116 |
| miR21
miR167 |
CHI
F3H |
Flavonoid biosynthesis
Anthocyanin accumulation |
117
|
| miR8181 | SPL | Flavonoids biosynthesis | 118 |
| miR14, miR22, miR31 | MYB33,WD 40 | Fruit development, Anthocyanin formation, hormone signalling | 119 |
Biological functions of flavonoids in plant defence
Responses against UV-B radiation
The expression of flavonoid genes is regulated by UV light120 which is evident from studies on various grass species growing in regions with high levels of solar UV-B rays contains high amount of flavonoids especially flavones (orientin and luteolin) as it provides protection against UV-B radiations121. Flavonoids are produced in response to exposure to UV-light and provide defence against UV stress response. This is due to the open planer structure of flavonoids that provides the UV-absorbing characteristics to these compounds122. The 3-OH group present in flavonoid skeleton chelates various metal ions, thus inhibiting the formation of free radicals and reactive oxygen species (ROS). UV absorbing properties of flavonoids have been intensively investigated in Arabidopsis, grapes and petunia.
Resistance against biotic and abiotic stresses
Secondary metabolites biosynthesis is also affected by environment which in turn influences the plant defence and pathogen attack123. Abiotic factors such as UV light regulates the flavonoid biosynthesis and also influences the response of plant to biotic stress like pathogen attack. In Arabidopsis, the production of flavonoids in response to both biotic (bacterial elicitor flg22) and abiotic stress (UB-B radiation) provides a structural barriers for pathogen spread. This is due to high transcript levels of flavonol synthase that provide resistance to biotic stresses124. Similarly in maize plant, the resistance to corn earworm (Helicoverpazea) is associated with high concentration of flavones thus providing protection125. Flavonoid genes are also involved in adaptation to abiotic stresses due to activating signal transduction pathway of various plant hormones. In Arabidopsis, MYB is the first gene reported to be produced during water stress that induces the abscisic acid biosynthesis pathway and results in closure of stomata, thus helping plant to combat water stress126,127. In barley, MYB gene was identified to be the first gene that mediates gibberellins signalling pathway, thus controlling plant growth and development128.
Roles of flavonoids in plant reproduction
Flavonoids imparts colour to the plant parts especially to pollens that aid in pollination and enhances plant reproduction129. The role of flavonoids in plant fertility was first studied in maize130 where it was observed that the mutants lacking chalcone synthase have non-functional pollen tube that leads to male sterility as well as flavonoid deficiency131. In tomato, parthenocarpy was induced due to silencing of chalcone synthase gene that resulted in production of seedless tomatoes132. Similarly, silencing of FLS gene resulted in production of non-functional pollen tube in tobacco plant and further implication of flavonol reversed the condition, suggesting its role in pollen fertility and its germination133.
Conclusion
Flavonoid metabolites play significant role in controlling the plant development and their interaction with various biotic and abiotic components. Due to the prevalence and beneficial health roles, flavonoid biosynthetic pathway has been intensively studied in many plants. Flavonoid biosynthesis and its accumulation is regulated in spatial and temporal manner and is also controlled by various genetic and development factors. About three decades of research is based on the conserved mechanism of flavonoid regulation that culminates in MBW complex, operating at transcriptional level controlling not only flavonoid pathway but also plant development pathway.
However, the underlying mechanisms that control the flavonoid regulation is still elusive and requires much investigation to dissect out the new perspective to understand the complexities involved in its regulation. The more in-depth study needs to be carried out to decipher the underlying mode of action, mode of intracellular transport, regulation and interaction of these flavonoid compounds in plants. This information would enable metabolic engineering of flavonoid pathway genes that can enhance their content, stability and bioactivity in plants in near future. Further, studies on various approaches such as constitutive over-expression or tissue specific expression aiming the miRNAs of interest and their corresponding target gene could be explored which could improve the response of plants to various biotic and abiotic stresses as well as their overall growth and development.
Acknowledgement
Fellowship from INSPIRE (DST/INSPIRE Fellowship/ 2013/1020), Department of Science and Technology, New Delhi is highly acknowledged. Authors are thankful to the School of Biotechnology, University of Jammu, Jammu for providing basic facilities.
Conflict of interest
The authors declare that they have no conflict of interest.
References
- Yonekura-Sakakibara K., Higashi Y., Nakabayashi R. The origin and evolution of plant flavonoid metabolism. Plant. Sci. 2019;doi: 10.3389/fpls. 2019.00943.
CrossRef - Kumar S., Pandey A. K. Chemistry and biological activities of flavonoids: an overview. World. J. 2013;162750.
CrossRef - Saito K., Yonekura-Sakakibara K., Nakabayashi R., Higashi Y., Yamazaki M., Tohge T., Fernie A. R. The flavonoid biosynthetic pathway in Arabidopsis: structural and genetic diversity. Physiol. Biochem. 2013;72:21-34.
CrossRef - Peer W. A., Murphy A. S. Flavonoids and auxin transport: modulators or regulators? Plant. Sci. 2007;12:556-63.
CrossRef - Tohge T., Fernie A. Specialized metabolites of the flavonol class mediate root phototropism and growth. Mol Plant. 2016;9:1554-55.
CrossRef - Winkel-Shirley B. Biosynthesis of flavonoids and effects of stress. Opin. Plant. Biol. 2002;5: 218-23.
CrossRef - Kelly E. H., Anthony R. T., Dennis J. B. Flavonoid antioxidants: chemistry, metabolism and structure-activity relationships. Nutr. Biochem. 2002;13:572-84.
CrossRef - Sharifi-Rad J., Quispe C., Shaheen S., El Haouari M., Azzini E., Butnariu M., Sarac I., Pentea M., Ramírez-Alarcón K., Martorell M., Kumar M., Docea A.O., Cruz-Martins N., Calina D. Flavonoids as potential anti-platelet aggregation agents: from biochemistry to health promoting abilities. Crit Rev Food Sci Nutr. 2021. doi: 10.1080/10408398.2021.1924612.
CrossRef - Petrache P., Rodino S., Butu M., Pribac G., Pentea M., Butnariu M. Polyacetylene and carotenes from Petroselinum sativum Dig J Nanomater Bios. 2014;9(4):1523-27.
- Dixon R. A., Paiva N. L. Stress-induced phenylpropanoid metabolism. Plant Cell. 1995;7:1085-97.
CrossRef - Wang H., Fan W., Li H., Yang J., Huang J., Zhang P. Functional characterization of dihydroflavonol-4-reductase in anthocyanin biosynthesis of purple sweet potato underlies the direct evidence of anthocyanins function against abiotic stresses. PLoS One. 2013;8:e78484.
CrossRef - Davies K. M., Schwinn K. E., Deroles S. C., Manson D. G., Lewis D. H., Bloor S. J., Bradley J. M. Enhancing anthocyanin production by altering competition for substrate between flavonol synthase and dihydroflavonol 4-reductase. Euphytica. 2002;131:259-68.
CrossRef - Barbat C., Rodino S., Petrache P., Butu M., Butnariu M. Microencapsulation of the allelochemical compounds and study of their release from different products. Dig J Nanomater Bios. 2013;8(3):945-53.
- Falcone Ferreyra M.L., Rius S.P., Casati P. Flavonoids: biosynthesis, biological functions, and biotechnological applications. Front Plant Sci. 2012;3:222. doi:10.3389/fpls.2012.00222.
CrossRef - Behrens C.E., Smith K.E., Iancu C.V., Choe J-N., Dean J.V. Transport of Anthocyanins and other Flavonoids by the Arabidopsis ATP-Binding Cassette Transporter AtABCC2. Sci Rep. 2019;9:437. https://doi.org/10.1038/s41598-018-37504-8.
CrossRef - Fu F., Zhang W., Li Y.Y., Wang H.L. Establishment of the model system between phytochemicals and gene expression profiles in Macrosclereid cells of Medicago truncatula.Sci Rep. 2017;7(1):2580.
CrossRef - Fu B., Ji X., Zhao M., He F., Wang X., Wang Y., Liu P., Niu L. The influence of light quality on the accumulation of flavonoids in tobacco (Nicotiana tabacum) leaves. J PhotochemPhotobiol B. 2016;162:544-549.
CrossRef - Bostan C., Butnariu M., Butu M., Ortan A., Butu A., Rodino S., Parvu C. Allelopathic effect of Festuca rubra on perennial grasses. Biotechnol. Lett.. 2013;18(2):8190-96.
- Mouradov A., Spangenberg G. Flavonoids: a metabolic network mediating plants adaptation to their real estate. Plant. Sci. 2014;5:620.
CrossRef - Butnariu M., Butu A. Chemical Composition of Vegetables and their Products. In: Cheung P. (eds) Handbook of Food Chemistry. Springer, Berlin, Heidelberg. 2014. https://doi.org/10.1007/978-3-642-41609-5_17-1
CrossRef - Butu M.,Rodino S., Butu A., Butnariu M. (2014). Screening of bioflavonoid and antioxidant activity of lens culinaris Dig J Nanomater Bios. 2014;9(2):519-29.
- Yang X. M., Jiang Y. M., Yang J. L., He J. R., Sun J., Chen F., Zhang M., Yang B. Prenylated flavonoids, promising nutraceuticals with impressive biological activities. Food. Sci. Technol. 2015;44:93-104.
CrossRef - Alzand K. I., Mohamed M. A. Flavonoids: Chemistry, biochemistry and antioxidant activity. Pharm. Res. 2012;5:4013-20.
- Kozlowski D., Trouillas P., Calliste C., Marsal P., Lazzaroni R., Duroux J. L. Density functional theory study of the conformational, electronic, and antioxidant properties of natural chalcones. Phys. Chem. A. 2007;111:1138-45.
CrossRef - Jandial D. D., Blair C. A., Zhang S., Krill L. S., Zhang Y. B., Zi X. Molecular targeted approaches to cancer therapy and prevention using chalcones. Cancer Drug Tar. 2014;14:181-200.
CrossRef - Rozmer Z., Perjési P. Naturally occurring chalcones and their biological activities. Rev. 2016;15:87-120.
CrossRef - Martens S., Preuss A., Matern U. Multifunctional flavonoid dioxygenases: flavonol and anthocyanin biosynthesis in Arabidopsis thaliana Phytochem. 2010;71:1040-49.
CrossRef - Gebhardt Y. H., Witte S., Steuber H., Matern U., Martens S. Evolution of flavone synthase I from parsley flavanone 3ß-hydroxylase by site-directed mutagenesis. Plant Physiol. 2007;144:1442-54.
CrossRef - Ververidis F., Trantas E., Douglas C., Vollmer G., Kretzschmar G., Panopoulos N. Biotechnology of flavonoids and other phenylpropanoid-derived natural products. Part II: reconstruction of multienzyme pathways in plants and microbes. J. 2007;2:1235-49.
CrossRef - Pervaiz T., Songtao J., Faghihi F., Haider M. S., Fang J. Naturally occurring anthocyanin, structure, functions and biosynthetic pathway in fruit plants. PlantBiochem. Physiol. 2017;doi:10.4172/2329-9029.1000187.
CrossRef - Choi J. Y., Lee S. J., Lee S. J., Park S., Lee J. H., Shim J. H., Abd El-Aty A. M., Jin J. S., Jeong E. D., Lee W. S., Shin S. C. Analysis and tentative structure elucidation of new anthocyanins in fruit peel of Vitis coignetiaePulliat (meoru) using LC‐MS/MS: Contribution to the overall antioxidant activity. Sep. Sci. 2010;33:1192-97.
CrossRef - Jaakola L. New insights into the regulation of anthocyanin biosynthesis in fruits. Trends Plant Sci. 2013;18:477-83.
CrossRef - Yang B., Liu H., Yang J., Gupta V. K., Jiang Y. New insights on bioactivities and biosynthesis of flavonoid glycosides. Trends Food Sci. Technol. 2018;79:116-24.
CrossRef - Roriz C. L., Barros L., Carvalho A. M., Santos-Buelga C., Ferreira I. C. F. R. Pterospartumtridentatum, Gomphrena globosa and Cymbopogon citratus: A phytochemical study focused on antioxidant compounds. Food Res. Int. 2014;64:684-93.
CrossRef - Frydman A., Weisshaus O., Bar-Peled M., Huhman D. V., Sumner L. W., Marin F. R., Lewinsohn E., Fluhr R., Gressel J., Eyal Y. Citrus fruit bitter flavors: isolation and functional characterization of the gene Cm1, 2RhaT encoding a 1,2 rhamnosyltransferase, a key enzyme in the biosynthesis of the bitter flavonoids of citrus. Plant J. 2004;40:88-100.
CrossRef - Pang Y., Peel G. J., Sharma S. B., Tang Y., Dixon R. A. A transcript profiling approach reveals an epicatechin-specific glucosyltransferase expressed in the seed coat of Medicago truncatula. Natl. Acad. Sci. 2008;105:14210-15.
CrossRef - Koornneef M. Mutations affecting the testacolor in Arabidopsis. Arabidopsis Information Service. 1990;28:1-4.
- Takos A. M., Jaffé F. W., Jacob S. R., Bogs J., Robinson S. P., Walker A. R. Light-induced expression of a MYB gene regulates anthocyanin biosynthesis in red apples. Plant Physiol. 2006;142:1216-32.
CrossRef - Grotewold E. The challenges of moving chemicals within and out of cells: insights into the transport of plant natural products. Planta.2004;219:906-9.
CrossRef - Koes R., Verweij W., Quattrocchio F. Flavonoids: a colorful model for the regulation and evolution of biochemical pathways. Trends Plant Sci. 2005;10:236-42.
CrossRef - Lin-Wang K., Bolitho K., Grafton K., Kortstee A., Karunairetnam S., McGhie T. K., Espley R. V., Hellens R. P., Allan A. C. An R2R3 MYB transcription factor associated with regulation of the anthocyanin biosynthetic pathway in Rosaceae. BMC Plant Biol. 2010;10:50.
CrossRef - Ferrer J. L., Jez J. M., Bowman M. E, Dixon R. A., Noel J. P. Structure of chalcone synthase and the molecular basis of plant polyketide biosynthesis. Struct. Biol. 1999;6:775-84.
- Ghanevati M., Jaworski J. G. Active-site residues of a plant membrane-bound fatty acid elongase β-ketoacyl-CoA synthase, FAE1 KCS. Biophys. Acta. 2001;1530:77-85.
CrossRef - Tropf S., Kärcher B., Schröder G., Schröder J. Reaction mechanisms of homodimeric plant polyketide synthase (stilbenes and chalcone synthase). A single active site for the condensing reaction is sufficient for synthesis of stilbenes, chalcones, and 6′-deoxychalcones. Biol. Chem. 1995;270:7922-28.
CrossRef - Dao T. T., Linthorst H. J., Verpoorte R. Chalcone synthase and its functions in plant resistance, In: Proceedings of the Phytochemical Society of Europe. Rev.2011;10 397-412.
CrossRef - Abe I., Morita H. Structure and function of the chalcone synthase superfamily of plant type III polyketide synthases. Prod. Rep. 2010;27:809-38.
CrossRef - Suh D. Y., Fukuma K., Kagami J., Yamazaki Y., Shibuya M., Ebizuka Y., Sankawa U. Identification of amino acid residues important in the cyclization reactions of chalcone and stilbene synthases. J. 2000;350:229-35.
CrossRef - Jez J. M., Noel J. P. Mechanism of chalcone synthase: pKa of the catalytic cysteine and the role of the conserved histidine in a plant polyketide synthase. Biol. Chem. 2000;275:39640-46.
CrossRef - Sun W., Meng X., Liang L., Jiang W., Huang Y., He J., Hu H., Almqvist J., Gao X., Wang L. Molecular and biochemical analysis of chalcone synthase from Freesiahybrid in flavonoid biosynthetic pathway. PLoS One. 2015;10:e0119054.
CrossRef - Van der Meer I. M., Spelt C. E., Mol J. N., Stuitje A. R. Promoter analysis of the chalcone synthase (chsA) gene of Petunia hybrida: A 67-bp promoter region directs flower-specific expression. Plant Mol. Biol. 1990;15:95-109.
CrossRef - Zheng D., Hrazdina G. Molecular and biochemical characterization of benzalacetone synthase and chalcone synthase genes and their proteins from raspberry (Rubus idaeus). Arch. Biochem. Biophys. 2008;470:139-45.
CrossRef - Tai D., Tian J., Zhang J., Song T., Yao Y. A Maluscrabapple chalcone synthase gene, McCHS, regulates red petal color and flavonoid biosynthesis. PLoS One. 2014;9:e0110570.
CrossRef - Wang C., Zhi S., Liu C., Xu F., Zhao A., Wang X., Tang X., Li Z., Huang P., Yu M. Isolation and characterization of a novel chalcone synthase gene family from mulberry. Plant Physiol. Biochem. 2017;115:107-18.
CrossRef - Jaakola L., Määttä K., Pirttilä A. M., Törrönen R., Kärenlampi S., Hohtola A. Expression of genes involved in anthocyanin biosynthesis in relation to anthocyanin, proanthocyanidin, and flavonol levels during bilberry fruit development. Plant Physiol. 2002;130:729-39.
CrossRef - Almeida J. R., D’Amico E., Preuss A., Carbone F., de Vos C. H., Deiml B., Mourgues F., Perrotta G., Fischer T. C., Bovy A. G., Matens S., Rosati C. Characterization of major enzymes and genes involved in flavonoid and proanthocyanidin biosynthesis during fruit development in strawberry (Fragaria x ananassa). Biochem. Biophys. 2007;465:61-71.
CrossRef - Yang Y. N., Yao G. F., Zheng D., Zhang S. L., Wang C., Zhang M. Y., Wu J. Expression differences of anthocyanin biosynthesis genes reveal regulation patterns for red pear coloration. Plant Cell Rep. 2015;34:189-98.
CrossRef - Kaur R., Aslam L., Kapoor N., Mahajan R. Identification and comparative expression analysis of chalcone synthase, flavanone 3‑hydroxylase and dihydroflavonol4‑reductase genes in wild pomegranate (Punica granatum) organs.Braz J Bot. 2020.https://doi.org/10.1007/s40415-020-00648-x.
CrossRef - Ngaki M. N., Louie G. V., Philippe R. N., Manning G., Pojer F., Bowman M. E., Li L., Larsen E., Wurtele E. S., Noel J. P. Evolution of the chalcone-isomerase fold from fatty-acid binding to stereospecific catalysis. Nature. 2012;485:530-33.
CrossRef - Li L. L., Cheng H., Xu F. Progress of chalcone isomerase in plants. Lett. 2008;19:935-37.
- Mehdy M. C., Lamb C. J. Chalcone isomerase cDNA cloning and mRNA induction by fungal elicitor, wounding and infection. EMBO J. 1988;86:1527-33.
CrossRef - Shimada N., Aoki T., Sato S., Nakamura Y., Tabata S., Ayabe S. A cluster of genes encodes the two types of chalcone isomerase involved in the biosynthesis of general flavonoids and legume-specific 5-deoxy (iso) flavonoids in Lotusjaponicas. Plant Physiol. 2003;131:941-51.
CrossRef - Jiang W., Yin Q., Wu R., Zheng G, Liu J., Dixon R. A., Pang Y. Role of a chalcone isomerase-like protein in flavonoid biosynthesis in Arabidopsis thaliana. Exp. Bot. 2015;66:7165-79.
CrossRef - He F., Pan Y. Purification and characterization of chalcone isomerase from fresh-cut Chinese water-chestnut. Food Sci. Technol. 2017;79:402-9.
CrossRef - Holton T. A., Cornish E. C. Genetics and biochemistry of anthocyanin biosynthesis. Plant Cell. 1995;7:1071-83.
CrossRef - Singh K., Rani A., Kumar S., Sood P., Mahajan M., Yadav S. K., Singh B., Ahuja P. S. An early gene of the flavonoid pathway, flavanone 3-hydroxylase, exhibits a positive relationship with the concentration of catechins in tea (Camellia sinensis). Tree Physiol. 2008;28:1349-56.
CrossRef - Kumar A., Singh B., Singh K. Functional characterization of flavanone 3-hydroxylase gene from Phyllanthus emblica (L.). Plant Biochem. Biotech. 2015;24:453-60.
CrossRef - Lukacin R ., Britsch L. Identification of strictly conserved histidine and arginine residues as part of the active site in Petunia hybrida flavanone 3 beta-hydroxylase. J. Biochem. 1997;249:748-57.
CrossRef - Williams R. J., Spencer J. P., Rice-Evans C. Flavonoids: antioxidants or signalling molecules? Free Radic. Biol. Med. 2004;36:838-49.
CrossRef - Sumalatha D. Antioxidant and Antitumor activity of Phyllanthus emblica in colon cancer cell lines. J. Curr. Microbiol. Appl. Sci. 2013;2:189-95.
- O’Reilly C., Shepherd N., Pereira A., Schwarz-Sommer Z., Bertram I., Peterson P. A., Saedler H. Molecular cloning of the a1 locus in Zea mays using the transposable elements En and Mu1. EMBO J. 1985;4: 877-82.
CrossRef - Jörnvall H., Persson B., Krook M., Atrian S., Gonzalez-Duarte R., Jeffery J., Ghosh D. Short-chain dehydrogenase/reductase (SDR). Biochem. 1995;34:6003-13.
CrossRef - Petit P., Granier T., d’Estaintot B. L., Manigand C., Bathany K., Schmitter J. M., Lauvergeat V., Hamdi S., Gallois B. Crystal structure of grape dihydroflavonol 4-reductase, a key enzyme in flavonoid biosynthesis. Mol. Biol. 2007;368:1345-57.
CrossRef - Johnson E. T., Ryu S., Yi H., Shin B., Cheong H., Choi G. Alteration of a single amino acid changes the substrate specificity of dihydroflavonol 4- reductase. Plant J. 2001;25:325-33.
CrossRef - Coutinho P. M., Deleury E., Davies G. J., Henrissat B. An evolving hierarchical family classification for glycosyltransferases. Mol. Biol. 2003;328:307-17.
CrossRef - Shao H., He X., Achnine L., Blount J. W., Dixon R. A., Wang X. Crystal structures of a multifunctional triterpene/flavonoid glycosyltransferase from Medicago truncatula. Plant Cell. 2005;17:3141-54.
CrossRef - Harborne J. B., Baxter H. The handbook of natural flavonoids (Wiley, New York, USA) 1999.
- Oancea S., Oprean L. Anthocyanins, from biosynthesis in plants to human health benefits. Food Technol. 2011;15:3-16.
- Kaur R., Aslam L., Kapoor N., Mahajan R. Molecular characterization of PgUFGT gene and R2R3-PgMYB transcription factor involved in flavonoid biosynthesis in four tissues of wild pomegranate (Punica granatum). J. Genet. 2019;98:94.
CrossRef - Das P. K., Shin D. H., Choi S. B., Park Y. I. Sugar-hormone cross-talk in anthocyanin biosynthesis. Mol Cells. 2012;34:501-7.
CrossRef - Chervin C., Tira-Umphon A., Chatelet P., Jauneau A., Boss P. K., Tesniere C. Ethylene and other stimuli affect expression of the UDP glucose-flavonoid 3-O-glucosyltransferase in a non-climacteric fruit. Vitis. 2009;48:11-16.
- Baudry A., Heim M. A., Dubreucq B., Caboche M., Weisshaar B., Lepiniec L. TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana. Plant J. 2004;39:366-80.
CrossRef - Patra B., Schluttenhofer C., Wu Y., Pattanaik S., Yuan L. Transcriptional regulation of secondary metabolite biosynthesis in plants. Biophys. Acta. 2013;1829:1236-47.
CrossRef - Xu W., Grain D., Bobet S., Le Gourrierec J., Thévenin J., Kelemen Z., Lepiniec L., Dubos C. Complexity and robustness of the flavonoid transcriptional regulatory network revealed by comprehensive analyses of MYB–bHLH–WDR complexes and their targets in Arabidopsis New Phytol. 2014;202:132-144.
CrossRef - Klempnauer K. H., Bonifer C., Sippel A. E. Identification and characterization of the protein encoded by the human c-myb protooncogene. EMBO J. 1986;5:1903-11.
CrossRef - Klempnauer K. H., Gonda T. J., Bishop J. M. Nucleotide sequence of the retroviral leukemia gene v-myb and its cellular progenitor c-myb: the architecture of a transduced oncogene. 1982;31:453-63.
CrossRef - Cone K. C., Burr F. A., Burr B. Molecular analysis of the maize anthocyanin regulatory locus C1. Natl. Acad. Sci. USA. 1986;83:9631-35.
CrossRef - Paz-Ares J., Ghosal D., Wienand U., Peterson P. A., Saedler H. The regulatory c1 locus of Zea mays encodes a protein with homology to myb proto-oncogene products and with structural similarities to transcriptional activators. EMBO J. 1987;6:3553-58.
CrossRef - Rosinski J. A., Atchley W. R. Molecular evolution of the Myb family of transcription factors: evidence for polyphyletic origin. Mol. Evol. 1998;46:74-83.
CrossRef - Jin H., Martin C. Multi-functionality and diversity within the plant MYB-gene family. Plant Mol. Biol. 1999;41:577-85.
CrossRef - Stracke R., Werber M., Weisshaar B. The R2R3-MYB gene family in Curr. Opin. Plant. Biol. 2001;4:447-56.
CrossRef - Spelt C., Quattrocchio F., Mol J. N., Koes R. Anthocyanin1 of petunia encodes a basic-helix loop helix protein that directly activates structural anthocyanin genes. Plant Cell. 2000;12:1619-31.
CrossRef - Schwinn K., Venail J., Shang Y., Mackay S., Alm V., Butelli E., Oyama R., Bailey P., Davies K., Martin C. A small family of MYB-regulatory genes controls floral pigmentation intensity and patterning in the genus Plant Cell. 2006;18:831-51.
CrossRef - Espley R. V., Hellens R. P., Putterill J., Stevenson D. E., Kutty-Amma S., Allan A. C. Red colouration in apple fruit is due to the activity of the MYB transcription factor, MdMYB10. Plant J. 2007;49: 414-27.
CrossRef - Dubos C., Stracke R., Grotewold E., Weisshaar B., Martin C., Lepiniec L. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010;15:573-81.
CrossRef - Ludwig S. R., Habera L. F., Dellaporta S. L., Wessler S. R. Lc, a member of the maize R gene family responsible for tissue-specific anthocyanin production, encodes a protein similar to transcriptional activators and contains the myc homology region. Natl. Acad. Sci. USA. 1989;86:7092-96.
CrossRef - Lloyd A., Brockman A., Aguirre L., Campbell A., Bean A., Cantero A., Gonzalez A. Advances in the MYB-bHLH-WD Repeat (MBW) pigment regulatory model: addition of a WRKY factor and co-option of an anthocyanin MYB for betalain regulation. Plant Cell Physiol. 2017;58:1431-41.
CrossRef - Feller A., Machemer K., Braun E. L., Grotewold E. Evolutionary and comparative analysis of MYB and bHLH plant transcription factors. Plant J. 2011;66:94-116.
CrossRef - Van Nocker S., Ludwig P. The WD-repeat protein superfamily in Arabidopsis: conservation and divergence in structure and function. BMC Genomics. 2003;4:50.
CrossRef - Khvorova A., Reynolds A., Jayasena S. D. Functional siRNAs and miRNAs exhibit strand bias. 2007;131:41-9.
- Brant E. J., Budak H. Plant small non-coding RNAs and their roles in biotic stresses. Plant Sci. 2018;doi: 10.3389/fpls.2018.01038.
CrossRef - Williams L., Grigg S. P., Xie M., Christensen S., Fletcher J. C. Regulation of Arabidopsis shoot apical meristem and lateral organ formation by microRNA miR166 g and its AtHD-ZIP target genes. Development. 2005;132:3657-68.
CrossRef - Sun C., Zhao Q., Liu D. D., You C. X., Hao Y. J. Ectopic expression of the apple Md-miRNA156 h gene regulates flower and fruit development in Plant Cell Tiss. Organ Cult. 2013;112:343-51.
CrossRef - Valencia-Sanchez M. A., Liu J., Hannon G. J., Parker R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 2006;20:515-24.
CrossRef - Shriram V., Kumar V., Devarumath R. M., Khare T. S., Wani S. H. miRNAs as potential targets for abiotic stress tolerance in plants. Plant Sci. 2016;7:817.
CrossRef - Hsieh L. C., Lin S. I., Shih A. C., Chen J. W., Lin W. Y., Tseng C. Y., Li W. H., Chiou T. J. Uncovering small RNA-mediated responses to phosphate deficiency in Arabidopsis by deep sequencing. Plant Physiol. 2009;151:2120-32.
CrossRef - Yang F., Cai J., Yang Y., Liu Z. Overexpression of microRNA828 reduces anthocyanin accumulation in Plant Cell Tiss. Organ Cult. 2013;115:159-67.
CrossRef - Jia X., Shen J., Liu H., Li F., Ding N., Gao C., Pattanaik S., Patra B., Li R., Yuan L. Small tandem target mimic-mediated blockage of microRNA858 induces anthocyanin accumulation in tomato. Planta. 2015;242:283-93.
CrossRef - Riese M., Hohmann S., Saedler H., Munster T., Huijser P. Comparative analysis of the SBP-box gene families in patens and seed plants. Gene. 2007;401:28-37.
CrossRef - Yu N., Cai W. J., Wang S., Shan C. M., Wang L. J., Chen X. Y. Temporal control of trichome distribution by microRNA156-targeted SPL genes in Arabidopsis thaliana. Plant Cell. 2010;22: 2322-35.
CrossRef - Wu G., Park M. Y., Conway S. R., Wang J. W., Weigel D., Poethig R. S. The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. 2009;138:750-59.
CrossRef - Gou J. Y., Felippes F. F., Liu C. J., Weigel D., Wang J. W. Negative regulation of anthocyanin biosynthesis in Arabidopsis by a miR156-targeted SPL transcription factor. Plant Cell. 2011;23: 1512-22.
CrossRef - Cui L. G., Shan J. X., Shi M., Gao J. P., Lin H. X. The miR156-SPL9-DFR pathway coordinates the relationship between development and abiotic stress tolerance in plants. Plant J. 2014;80:1108-17.
CrossRef - Chai J., Feng R., Shi H., Ren M., Zhang Y., Wang J. Bioinformatic identification and expression analysis of banana microRNAs and their targets. PLoS One. 2015;10:e0123083.
CrossRef - Li Y., Cui W., Wang R., Lin M., Zhong Y., Sun L., Qi X., Fang J. MicroRNA858-mediated regulation of anthocyanin biosynthesis in kiwifruit (Actinidia arguta) based on small RNA sequencing.PLoS ONE. 2019;14:e0217480.
CrossRef - Gupta O. P., Suhaset G. K., Sagar B., Meena N. L., Dahuja A. Contemporary understanding of miRNA-based regulation of secondary metabolites biosynthesis in plants. Plant Sci. 2017;8: 374.
CrossRef - Kim J., Park J. H., Lim C. J., Lim J. Y., Ryu J. Y., Lee B. W., Choi J. P., Kim W. B., Lee H. Y., Choi Y., Kim D., Hur C. G., Kim S., Noh Y. S., Shin C., Kwon S. Y. Small RNA and transcriptome deep sequencing proffers insight into floral gene regulation in Rosa BMC Genomics. 2012;13: 657.
CrossRef - Yue J. Y., Lu X. H., Zhang H., Ge J., Gao X., Liu Y. Identification of conserved and novel microRNAs in blueberry. Plant Sci. 2017;8:1155.
CrossRef - Sun Y., Qiu Y., Duan M., Wang J., Zhang X., Wang H., Song J., Li X. Identification of anthocyanin biosynthesis related microRNAs in a distinctive chinese radish (Raphanus sativus) by high throughput sequencing. Mol. Gene Genomics. 2016;292:1-15.
CrossRef - Saminathan T., Bodunrin A., Singh N. V., Devarajan R., Nimmakayala P., Jeff M., Aradhya M., Reddy U. K. Genome-wide identification of microRNAs in pomegranate (Punica granatum) by high-throughput sequencing. BMC Plant Biol. 2016;16:122.
CrossRef - Zoratti L., Karppinen K., Escobar A. L., Häggman H., Jaakola L. Light-controlled flavonoid biosynthesis in fruits. Plant Sci. 2014;5:534.
CrossRef - Van De Staaij J., De Bakker N. V., Oosthoek A., Broekman R., Van Beem A., Stroetenga M., Aerts R., Rozema J. Flavonoid concentrations in three grass species and a sedge grown in the field and under controlled environment conditions in response to enhanced UV-B radiation. PhotochemPhotobiol. 2002;66:21-9.
CrossRef - Agati G., Brunetti C., Di Ferdinando M., Ferrini F., Pollastri S., Tattini M. Functional roles of flavonoids in photoprotection: New evidence, lessons from the past. Plant Physiol. Biochem. 2013;72:1-11.
CrossRef - Yang L., Wen K. S., Ruan X., Zhao Y. X., Wei F., Wang Q. Response of plant secondary metabolites to environmental factors. 2018;doi: 10.3390/ molecules23040762.
- Schenke D., Böttcher C., Scheel D. Crosstalk between abiotic ultraviolet-B stress and biotic (flg22) stress signalling in Arabidopsis prevents flavonol accumulation in favor of pathogen defence compound production. Plant Cell Environ. 2011;34:1849-64.
CrossRef - Johnson E. T., Berhow M. A., Dowd P. F. Expression of a maize myb transcription factor driven by a putative silk-specific promoter significantly enhances resistance to Helicoverpazea in transgenic maize. Agr. Food Chem. 2007;55:2998-3003.
CrossRef - Abe H., Yamaguchi-Shinozaki K., Urao T., lwasaki T., Hosokawa D., Shinozaki K. Role of Arabidopsis MYC and MYB homologs in drought and abscisic acid-regulated gene expression. Plant Cell. 1997;9:1859-68.
CrossRef - Maeda K., Kimura S., Demura T., Takeda J., Ozeki Y. DcMYB1 acts as a transcriptional activator of the carrot phenylalanine ammonia-lyase gene (DcPAL1) in response to elicitor treatment, UV-B irradiation and the dilution effect. Plant Mol. Biol. 2005;59:739-52.
CrossRef - Du H., Zhang L., Liu L., Tang X. F., Yang W. J., Wu Y. M., Huang Y. B., Tang Y. X. Biochemical and molecular characterization of plant MYB transcription factor family. 2009;74:1-11.
CrossRef - Van Der Meer I. M., Stam M. E., Van Tunen A. J., Mol J. N., Stuitje A. R. Antisense inhibition of flavonoid biosynthesis in petunia anthers results in male sterility. Plant Cell. 1992;4:253-62.
CrossRef - Mo Y., Nagel C., Taylor L. P. Biochemical complementation of chalcone synthase mutants defines a role for flavonols in functional pollen. Natl. Acad. Sci. USA. 1992;89:7213-17.
CrossRef - Pollak P. E., Vogt T., Mo Y., Taylor L. P. Chalcone synthase and flavonol accumulation in stigmas and anthers of Petunia hybrid.Plant Physiol. 1993;102:925-32.
CrossRef - Schijlen E. G., De Vos C. H., Martens S., Jonker H. H., Rosin F. M., Molthoff J. W., Tikunov Y. M., Angenent G. C., Van Tunen A. J., Bovy A. G. RNA interference silencing of chalcone synthase, the first step in the flavonoid biosynthesis pathway, leads to parthenocarpic tomato fruits. Plant Physiol. 2007;144:1520-30
CrossRef - Mahajan M., Ahuja P. S., Yadav S. K. Post-transcriptional silencing of flavonol synthase mRNA in tobacco leads to fruits with arrested seed set. PLoS ONE. 2011;6:e28315.
CrossRef

This work is licensed under a Creative Commons Attribution 4.0 International License.






