Manuscript accepted on : 21-Sep-19
Published online on: 30-09-2019
Plagiarism Check: Yes
Reviewed by: Graham Clark
Second Review by: Agata Piestrzyńska-Kajtoch
A New PCR-Based Species Genotyping Differentiation Approach in Entamoeaba
Dalia Abuljadayel1, Ahmed Atef1, Mohammed Al-Matary1, Sherif Edris1,2,3 
![]() , Khalid M. Al-Ghamdi1
, Khalid M. Al-Ghamdi1 , Nahid H. Hajrah1, Jamal S.M. Sabir1
, Nahid H. Hajrah1, Jamal S.M. Sabir1  , Neil Hall1,4 and Ahmed Bahieldin1,2*
, Neil Hall1,4 and Ahmed Bahieldin1,2*
1Department of Biological Sciences, Faculty of Science, King Abdulaziz University (KAU), P.O. Box 80141, Jeddah 21589, Saudi Arabia
2Department of Genetics, Faculty of Agriculture, Ain Shams University, Cairo, Egypt
3Princess Al-Jawhara Al-Brahim Centre of Excellence in Research of Hereditary Disorders (PACER-HD), Faculty of Medicine, King Abdulaziz University (KAU), Jeddah, Saudi Arabia
4The Genome Analysis Center, Norwich Research Park, Norwich, NR4 7UH, UK
Corresponding Author E-mail: abmahmed@kau.edu.sa
DOI : http://dx.doi.org/10.13005/bbra/2764
ABSTRACT: The most commonly used approach for Entamoeba species differentiation up to date is the tRNA-linked STR regions of the parasite’s genome. In the present study, a new reliable, fast and easy molecular tool for species differentiation was developed. DNA was isolated from fecal samples collected from infected subjects with either Entamoeba histolytica (EH) or Entamoeba disper (ED) in Saudi Arabia. Two types of primer sets were compared in which the first targeted tRNA-linked STR regions, while the second was designed after multiple contig alignment of the two genomes using NUCmer program in aligned areas with high similarity (~90%) and difference between of ~90 bp. The selection criteria secures that designed primers should pair with both EH and ED contig sequences at homologous regions of 200-500 bp of both species except for the presence of indels that result in the recovery of amplicons of two species with different sizes. Banding patterns in the tRNA-linked STR region resulted in the occurrence of several common amplicons. We speculate that primers mismatch with regions other than the specified STR arrays of Entamoeba histolytica or Entamoeba disper with organisms other than Entamoeba existed in the fecal sample. However, the STR-based approach looked very useful in studying strain differentiation and parasite diversity. The results for the new approach complemented those of the STR-based approach, except that the latter failed to detect coinfected subjects. The new approach proved to be useful at the species level, while the tRNA-linked STR approach can still be a good choice for strain differentiation.
KEYWORDS: Amplicon; Contig; Nucmer; pecies differentiation; STR; Strain Differentiation
Download this article as:| Copy the following to cite this article: Abuljadayel D, Atef A, Al-Matary M, Edris S, Al-Ghamdi k. M, Hajrah N. H, Sabir J. S. M, Hall N, Bahieldin A. A New PCR-Based Species Genotyping Differentiation Approach in Entamoeaba. Biosci Biotech Res Asia 2019;16(3). |
| Copy the following to cite this URL: Abuljadayel D, Atef A, Al-Matary M, Edris S, Al-Ghamdi k. M, Hajrah N. H, Sabir J. S. M, Hall N, Bahieldin A. A New PCR-Based Species Genotyping Differentiation Approach in Entamoeaba. Biosci Biotech Res Asia 2019;16(3). Available from: https://www.biotech-asia.org/?p=34206 |
Introduction
Amebiasis is a disease basically caused by the pseudopod-forming protozoan parasite Entamoeba histolytica (EH). The disease can either be asymptomatic [1] or can result in severe infection with amebic colitis (AC) and amebic liver abscess (ALA). AC is the main cause of diarrhea worldwide for children up to two years old especially those living in the rural developing countries. The disease represents the third leading cause of death, accounting for 9% of all deaths in children up to five years of age [2-4]. In Saudi Arabia, there are several communicable diseases affecting the human digestive system likely associated with Hajj season. EH is likely to be one of the most common causes of infectious diarrhea among Hajji individuals in addition to the diseases transferred from individuals coming from endemic areas [5]. The main problem lies in the possible transmission of new invasive strains of the parasite during this season.
Previous efforts of differentiation at the microscopic level failed to discriminate among Entamoeba species. Advances in molecular diagnostic methodologies have resulted in the recognition and separation of EH from the three other nonpathogenic species that infect humans. These other species are morphologically indistinguishable from EH. They are E. moshkovskii that may cause diarrhea [6], in addition to the nonpathogenic E. dispar (ED) and the newly described E. Bangladeshi [7-9]. This scenario changed when ED strains were lately isolated from symptomatic patients in Brazil [10]. These ED strains were able to cause AC and ALA that are occasionally indistinguishable from those produced by EH. This finding revived the possibility that ED can produce lesions in humans.
As ED was reported to be several times more common than EH worldwide [4], it is a must to detect discrete characteristics of this species as compared to EH. However, as indicated earlier, discrimination between Entamoeba histolytica (EH) and Entamoeba dispar (ED) as two separate species is still a difficult task. As a more complication, not all EH infections lead to disease in the host as only ~10% infections progresses to the development of clinical symptoms [6]. These findings indicate that the outcome of EH infection is still a mystery but we speculate it is strain-specific. In addition, evidences of coinfection with EH and ED was reported in some areas of endemicity with both parasites [11]. The later poses more burden into the characterization of either species, a phenomenon that exists in our study.
Molecular characterization based on the short tandem repeats (STRs) linked to tRNA genes of either species indicates the existence of a large number of subspecies or strains [12]. The latter type of differentiation, e.g., STRs, poses more burden in detecting a certain marker with a specific molecular weight for either species, on one hand, in addition to the possibility to get successive PCR products with similar sizes in other Entamoaba or non-Entamoeba species, on the other hand. Therefore, we found it useful to develop a reliable, fast and easy molecular tool for species differentiation, while recommend the use of STR markers only for strain differentiation. The first approach requires studying no more than one locus, while the second requires studying several loci for discrete strain differentiation [11,13].
Materials and Methods
Sample collection and DNA isolation
Fecal samples were collected from 37 dysentery-infected subjects, either Saudi or non-Saudi, in four hospitals in Jeddah, KAU. An ethical approval (no. A00451) has been issued by the Ministry of Health, Saudi Arabia following the regulation of the General Administration for Research and Studies at the Ministry of Health (registration no. 1195437) and the National Committee for Medical and Biological Ethics (registration no. H-02-J-002). Consent forms were filled by infected subjects or their relatives at sampling time.
The fecal samples used for molecular characterization were kept fresh at 4°C and diagnosed by microscopic examination and positive samples were subjected to DNA extraction by using QIAMP mini kit specific for stool purification (QIAamp® DNA Mini kit, Qiagen GmbH, Hilden, Germany) following manufacturer’s protocol. In order to remove RNA contamination, RNase A (10 mg/ml, Sigma, USA) was further used to DNA samples and incubated at 37oC for 30 min. The purity and concentration of DNA in the extracts were checked by the nanodrop (NanoDrop 2000 spectrophotometer, Thermo Scientific™, Thermo-Fisher scientific, DE, USA).
Primers used and PCR conditions
Two types of primers were used in the present study. The first type of primers (either genus- or species-specific) were originally designed by Ali et al. [11] in the tRNA-linked STR regions to amplify those of the EH HM-1:IMSS tRNA gene sequences (GenBank accession numbers BK005648-BK005672). Genus-specific primers of tRNA-linked STR of RTCT and NK1 arrays were used to amplify both EH and ED DNAs, while species-specific primers of tRNA-linked STR of RTCT were designed to amplify DNAs from one species only (Table 1 & Figure S1). Nomenclature of these strain differentiation primers derived from the single-letter amino acid code for the relevant tRNA genes flanking the STRs being amplified. Consensus array unit sequences and STR organizations are shown in the link http://homepages.lshtm.ac.uk/entamoeba/units/units.htm. Accession nos. used in designing primers of the RTCT and NK1 tRNA-linked STR arrays are BK005654.1 and BK005655.1 for EH, while HQ439972.1 and EF421344.1 for ED, respectively (Figure S1). PCR was performed using a ready-to-use master mix (BioTaq Green Master Mix, Promega) with DNA concentration of ~50 ng and conditions were 95ºC/5 min (initial denaturation), 95ºC/30 sec, 55-58ºC/45 sec and 72ºC/1 min (36 cycles), 72ºC (final extension), then reaction was held at 4ºC. Amplicons were originally run on agarose gel (1.5% in 1x TBE buffer) and successful amplicons were run using polyacrylamide gel electrophoresis (PAGE) [14] to recover amplicons with higher resolution. Either gel type was stained with ethidium bromide (0.3 ug/ml), then visually examined with UV transilluminator and photographed using a CCD camera (UVP, Cambridge, UK).
 |
Figure S1: Sequences of the RTCT (a & b) and NK1 (c & d) tRNA arrays of the EH species with accession nos. BK005654.1 (a) and BK005655.1 (c) and ED species with accession nos. |
Table 1: Primers generated used for strain differentiation.
| tRNA primers | Sequence (5` to 3`) | Annealing
temp. (°C) |
| General primers | ||
| R-R5
R-R3 |
AGCATCAGCCTTCTAAGCTG
CTTCCGACTGAGCTAACAAG |
55 |
| N-K5
N-K3 |
CGAACGGCTGTTAACCGTTA
TTCCTAGCTCAGTCGGTAGA |
55 |
| EH -specific primers | ||
| RR-H5
RR-H3 |
GCGCCTTTTTATTCAATATACTCC
GGATGAAGATATCTTCACAGGG |
57 |
| ED -specific primers | ||
| RR-D5
RR-D3 |
CATGAGGCGCCTTTTTATCA
AGGGATGATGATATTGAACACACTC |
58 |
In order to design the new species-specific primers, genomes of EH (https://www.ncbi.nlm.nih.gov/genome/27) and ED (https://www.ncbi.nlm.nih.gov/genome/372) were retrieved from NCBI (https://www.ncbi.nlm.nih.gov/genome/?term=entamoeba). Contigs of the two genomes (https://www.ncbi.nlm.nih.gov/Traces/wgs/AAFB02?display=contigs, https://www.ncbi.nlm.nih.gov/Traces/wgs/AANV02?display=contigs) were downloaded from NCBI. Multiple contig alignment of the two genomes was done using NUCmer module 3.0 (NUCleotide MUMmer, part of mummer software) to determine the position and orientation of a set of sequence contigs, and to find all of the maximal unique matches of a given length between the two input sequences, a step to increase the overall coverage of the alignment [15]. Only about 100 contigs of each species showed partial matching in DNA sequences. Commands were made to recover a delta file, which was converted to a coords file. The latter file type is accessible by Excel (xlxs). The contig pairs with high similarity (~90%) and difference between aligned area with ~90 bp (sequence similarity/difference criteria) were selected in the recovered Excel file. Different groups of primers were detected in this study for contig pairs meeting the sequence similarity/difference criteria. PCR was performed using ready master mix (BioTaq Green Master Mix, Promega) and conditions were 95ºC/5 min (initial denaturation), 95ºC/30 sec, 52ºC/45 sec and 72ºC/1 min (40 cycles), 72ºC (final extension), then reaction was held at 4ºC. Amplicons were run on agarose gel, stained with ethidium bromide (0.3 ug/ml), then visually examined and photographed.
Results
A number of 37 subjects (19 males and 18 females) aged between 1 and ˃ 50 years with background from more than seven countries participated in the study (Table 2). Saudi individuals (~66%) and those of 1-10 years old (~46%) represented the most frequent subjects of the two categories (e.g., nationality and age) in the study. Double survey of infection in tRNA-linked STR arrays indicated that all these subjects are infected with Entamoeba (Figures 1 & 2). All types of tRNA-linked STRs with Entamoeba general primers generated by Ali et al. [11] indicated that only two arrays, e.g., RTCT and NK1, out of the six arrays of STRs were successful in detecting the disease in the respective subjects (Figures 1 & 2). Gradient PCRs for either array (e.g., RTCT or NK1) were done in the present study to detect the best PCR thermal conditions and avoid the occurrence of false positives. The results indicated that annealing temperature previously indicated by Ali et al. [11] is the best.
Table 2: Detailed sociodemographic characteristics of the infected subjects living in Jeddah, Saudi Arabia.
| Serial no. | Sex | Nationality | Age (years old) | Species | Overall numbers | |
| 1 | Male | Syrian | 1-10 | ED | Gender | |
| 2 | Saudi | Male | 19 | |||
| 3 | Female | 18 | ||||
| 4 | EH | Age (years old) | ||||
| 5 | Yamani | 1-10 | 17 | |||
| 6 | Saudi | 11-20 | 1 | |||
| 7 | Yamani | 21-30 | ED | 21-30 | 7 | |
| 8 | EH | 31-40 | 7 | |||
| 9 | Saudi | 41-50 | 3 | |||
| 10 | Pakistani | >50 | 2 | |||
| 11 | Egyptian | 31-40 | ED | Nationality | ||
| 12 | Saudi | Saudi | 22 | |||
| 13 | Egyptian | 5 | ||||
| 14 | Egyptian | Yemeni | 5 | |||
| 15 | EH | Pakistani | 2 | |||
| 16 | Saudi | Other | 3 | |||
| 17 | Bangladesh | 41-50 | ED | Infecting species | ||
| 18 | Saudi | EH | EH | 22 | ||
| 19 | >50 | ED | 12 | |||
| 20 | Female | Saudi | 1-10 | ED | EH/ED | 3 |
| 21 | ||||||
| 22 | Yamani | EH | ||||
| 23 | ||||||
| 24 | Philippian | |||||
| 25 | Saudi | |||||
| 26 | ED | |||||
| 27 | EH | |||||
| 28 | ||||||
| 29 | ||||||
| 30 | ||||||
| 31 | 11-20 | ED | ||||
| 32 | Pakistani | 21-30 | EH | |||
| 33 | Saudi | |||||
| 34 | ||||||
| 35 | 31-40 | ED | ||||
| 36 | Egyptian | 41-50 | EH | |||
| 37 | >50 | |||||
Percentage of non-Saudi male subjects is 47.4% as compared to Saudi (52.6%)
Percentage of non-Saudi female subjects is 33.3% as compared to Saudi (66.7%)
Analysis of STR markers with Entamoeba general primers
The expected number and sizes of amplicons to be generated by both types of markers, either general or species-specific, are shown in Table S1. For the general markers, numbers of three and six amplicons in RTCT array of EH (e.g., 678, 686, 694 bp) and ED (e.g., 586, 686, 696, 702, 712, 728 bp), and numbers of two and one amplicons in NK1 array of EH (e.g., 527, 598 bp) and ED (e.g., 597 bp), respectively, were generated in Entamoeba sp. strains available in the NCBI (https://blast.ncbi.nlm.nih.gov/). This data scopes the light on the divergence of Entamoeba sp. strains in the linked-tRNA STR arrays. As our subjects are from different geographic backgrounds, we expected to detect several new strains in the studied subjects.
Banding patterns of either marker in the present study indicated the occurrence of several common and specific amplicons (Figures 1 & 2). We can simply explain the occurrence of specific amplicons that indicate the existence of new strains but we found it difficult to explain the occurrence of several common bands except that the two sets of primers mismatch with regions other than the two specified arrays in Entamoeba species or in organisms other than Entamoeba that surely exist in the fecal sample. Results of the RTCT array in the present study indicated the existence of three strain-specific markers in subjects 3 (>900 bp), 19 (~380 bp) and 24 (~400 bp). Amplicon sizes of these possibly new strains match none of those available in the NCBI link. The first two subjects are Saudi aged 1-10 and >50 years old, while the third is Philippian aged 1-10 years old (Figure 1). Results of the NK1 array indicated the existence of four strain-specific markers in subjects 6 (~1500 bp), 9 (~180 bp), 11 (~50 bp) and 32 (~160 bp). The first two subjects are Saudi aged 1-10 and 21-30 years old, while the third is Egyptian aged 31-40 years old and the forth is Pakistani aged 21-30 years old (Figure 2).
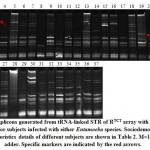 |
Figure 1: Amplicons generated from tRNA-linked STR of RTCT array with R-R5/R-R3 primers for subjects infected with either Entamoeba species. |
 |
Figure 2: Amplicons generated from tRNA-linked STR of NK1 array with N-K5/N-K3 primers for subjects infected with either Entamoeba species. Sociodemographic characteristics details of different subjects are shown in Table 2. M=100 bp ladder. Specific markers are indicated by the red arrows |
Analysis of STR markers with Entamoeba species-specific primers
For the species-specific markers in Table S1, numbers of nine and six amplicons in RTCT array of EH (e.g., 477, 485, 557, 565, 573, 581, 589, 597, 605 bp) and ED (e.g., 272, 598, 614, 630, 634, 646 bp), respectively, were generated in Entamoeba sp. strains available in the NCBI (https://blast.ncbi.nlm.nih.gov/). Based on the studied tRNA-linked STR of RTCT with species-specific primers (e.g., EH or ED) in the present study, 14 out of all subjects interestingly infected with symptomic ED strain(s). This number represents ~38% of the studied cases (Table 2).
 |
Table S1: Primer pairs generated to amplify short tandem repeats (STRs) regions of the tRNA arrays either in Entamoeba genus (general primers, e.g., R-R5/R-R3, N-K5/N-K3) or in a given Entamoeba species |
Results of the RTCT array of EH-infected subjects indicated the possible existence of two strain-specific markers in subjects 18 (>180 bp) and 24 (~400 bp) (Figure 3). The latter marker was detected for the same subject (Philippian) using general primer of the same array (Figure 1), while the first marker was generated for a Saudi subject aged 31-40 years old (Figure 3). Amplicon sizes of these possibly new strains match none of the nine EH strains available in the NCBI link. On the other hand, results of RTCT array in ED-infected subjects indicated the possible existence of as high as 10 new strain-specific markers in eight subjects (Figure 4). Marker sizes are 900 bp for subject 1, 200 bp for subject 2, 1300 bp for subject 3, 380 bp for subject 11, 200 bp for subject 12, 1400 and 280 bp for subject 14, 350 bp for subject 20 and 100 and 120 bp for subject 26. Two of these subjects are Egyptian (11 and 14), while one (1) is Syrian, while the other six subjects are Saudi (Table 2). Six of these markers are for subjects aged 1-10 years old, while four markers are for subject aged 31-40 years old (Figure 4). Markers of these possibly new strains match none of those available in the NCBI link, except for the marker sized ~280 bp of the Egyptian subject 14 that matches the size of the marker of ED isolate PRIR1 (272 bp) available in the NCBI link.
 |
Figure 3: Amplicons generated from tRNA-linked STRs with RR-H5/RR-H3 primers for subjects infected with EH. Sociodemographic characteristics details of different subjects are shown in Table 2. G=Gender, A=age (in years), N=Nationality, S=Saudi, E=Egyptian, Y=Yemeni, Pa=Pakistan, O=others. M=100 bp ladder. Specific markers are indicated by the red arrows. |
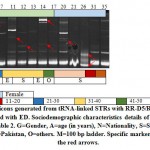 |
Figure 4: Amplicons generated from tRNA-linked STRs with RR-D5/RR-D3 primers for subjects infected with ED. Sociodemographic characteristics details of different subjects are shown in Table 2. G=Gender, A=age (in years), N=Nationality, S=Saudi, E=Egyptian, Y=Yemeni, Pa=Pakistan, O=others. M=100 bp ladder. Specific markers are indicated by the red arrows. |
It is likely to detect Entamoeba strains with specific markers in Egyptian (four markers), Syrian (one marker), Philippian (one marker), and Pakistani (one marker) individuals, but this is unlikely for Saudi individuals unless they were originally infected while they are abroad. Egyptian individuals are strong candidate of disease transmission to the Saudi habitat. Possible new strain-specific markers frequently found for subjects aged 1-10 years old across nationalities. Overall data indicate the possible occurrence of new strains in 14 out of the 37 subjects of which 10 of them are males (subjects 1, 2, 3, 6, 9, 11, 12, 14, 18, and 19), while four are females (subjects 20, 24, 26 and 32). This indicates that males are higher transmitter of the new strains than female.
Analysis of the new approach of species differentiation in Entamoeba
The new type of species-specific markers was developed to overcome the complications of the STR-based species differentiation method. In addition, we speculate that primers used for the tRNA-linked STR markers might mismatch with organisms other than Entamoeba existing in the fecal sample. However, we have followed an approach that is not devoid the same assumption. Therefore, we have developed five sets of primer pairs utilizing our new approach in order to choose the one that clearly matches with both Entamoeba species with no other amplicon’s background.
In the new approach, genomes of EH and ED were retrieved from NCBI (https://www.ncbi.nlm.nih.gov/genome/?term=entamoeba). Multiple contig alignment of the two genomes was done using NUCmer module (part of mummer 3.0 software, http://mummer.sourceforge.net/). This module is the most user-friendly alignment script for standard DNA sequence alignment. It is used to determine the position and orientation of a set of sequence contigs in relation to a finished sequence. The program is a three-step process comprising maximal exact matching, match clustering, and alignment extension. It searches the maximal unique matches of a given length between the two input sequences. Then, individual matches are clustered into closely grouped sets and the non-exact sequence between matches is aligned via a modified Smith-Waterman algorithm.
Commands were made to recover a delta file, which is converted to a coords file. The latter is accessible by Excel (xlxs). As outputs of the analysis, [S1] is the start of the alignment region, while [E1] is the end of the alignment region in the ED contig. [S2] is the start of the alignment region, while [E2] is the end of the alignment region of EH contig. [LEN 1] is the length of the alignment region in the ED contig, while [LEN 2] is the length of the alignment region in the EH contig. [% IDY] is the percent identity of the alignment. [LEN R] is the length of the ED contig, while [LEN Q] is the length of the EH contig. [COV R] is the percent alignment coverage in the ED contig, while [COV Q] is the percent alignment coverage in the EH contig.
Table S2 indicates the contig pairs of ED and EH ~90 bp difference and similarity between aligned area of ~90% were selected in the recovered Excel file. The selection criteria secures that species-specific primers to be designed should pair with both EH and ED contig sequences at completely homologous regions of 200-500 bp of both species except for the presence of indels that result in the amplification of DNA fragments of the two species with difference in sequence length. Five candidate contig pairs of ED and EH were found suitable for primer design in which each primer pair can amplify different product sizes for the two species (Table 3). Primers generated from these five pairs of contigs are shown in Table 4. A model of the alignment is shown in Table S3 and described in Figure 5 of which two contigs of ED (NW_001855238.1) and EH (NW_001915012.1) showed three aligned regions with size differences of 28, 58 and 126 bp. The region with the largest difference, e.g., 126 bp, was selected for primer design. In the model case, a difference of 45 bp length was recovered between amplicons of the two species (Table S3 & Figure 5).
Table S3: Statistics of paired alignment of two selected contigs of the two genomes of ED (NW_001855238.1) and EH (NW_001915012.1) using Nucmer module (NUCleotide MUMmer, part of mummer software). The contig pair showed high similarity (~90%) in three areas with ~4900, 4000 and 5700 bp total size. The difference in sizes of the three areas of the two contigs are 58, 126 and 28 bp, respectively. The area with 4900 bp with the largest difference of 126 bp, then the region within this area with the longest indel of 45 bp were selected. Sequences flanking this indel was utilized in designing primers for species-specific marker detection to generate 217 and 262 bp for the ED and EH contigs, respectively.
| S1 | E1 | length R (ED) | S2 | E2 | length Q (EH) | difference (ED-EH) | % IDY | [LEN R] [LEN Q] | [COV R] [COV Q] | contig no. (ED) | contig no. (EH) |
| 10179 | 15042 | 4863 | 27301 | 22380 | -4921 | 58 | 89.41 | 41758 39477 | 11.65 12.47 | NW_001855238.1 | NW_001915012.1 |
| 15513 | 19503 | 3990 | 21944 | 17828 | -4116 | 126 | 87.11 | 41758 39477 | 9.56 10.43 | NW_001855238.1 | NW_001915012.1 |
| 19774 | 25478 | 5704 | 17692 | 11960 | -5732 | 28 | 91.04 | 41758 39477 | 13.66 14.52 | NW_001855238.1 | NW_001915012.1 |
[S1] start of the alignment region in the reference sequence (ED)
[E1] end of the alignment region in the reference sequence (ED)
[S2] start of the alignment region in the query sequence (EH)
[E2] end of the alignment region in the query sequence (EH)
[LEN 1] length of the alignment region in the reference sequence (ED)
[LEN 2] length of the alignment region in the query sequence (ED)
[% IDY] percent identity of the alignment (ED/EH)
[LEN R] length of the reference sequence (ED)
[LEN Q] length of the query sequence (EH)
[COV R] percent alignment coverage in the reference sequence (ED)
[COV Q] percent alignment coverage in the query sequence (EH)
 |
Figure 5: A model for the approach used in selecting contigs of the ED and EH genomes for generating the new type of species-specific markers. |
Table 3: Contigs of EH and ED meeting the criteria of selecting contig pairs from multiple sequence alignment of contigs of the two genomes using nucmer module (part of mummer software).
| Contig pair no. | Species | Contig accession no. | Contig length (bp) | Diff. (bp) | Similarity
(%) |
| 1 | ED | NW_001854967.1 | 13230 | 128 | 91.88 |
| EH | NW_001914891.1 | 13102 | |||
| 2 | ED | NW_001855840.1 | 12450 | 180 | 91.14 |
| EH | NW_001915091.1 | 12630 | |||
| 3 | ED | NW_001853623.1 | 3840 | 111 | 89.99 |
| EH | NW_001915426.1 | 3951 | |||
| 4 | ED | NW_001854085.1 | 5132 | 107 | 90.01 |
| EH | NW_001915137.1 | 5025 | |||
| 5 | ED | NW_001855238.1 | 3991 | 126 | 87.11 |
| EH | NW_001915012.1 | 4117 |
Table 4: Primers designed based on contig pair analysis of EH and ED meeting the criteria of selecting contig pairs from multiple contig alignment of the EH and ED genomes using nucmer
| Contig pair no. | Name | Sequence | Product size (bp) | |
| ED | EH | |||
| 1 | P1_HD-F | AAAwCTTTCTTyrACTTCTTCTTCC | 424 | 531 |
| P1_HD-R | TTTAGGTTTTTCAGTTGCCAATC | |||
| 2 | P2_HD-F | CATTGACTTTCAGGAGGrAATTG | 423 | 476 |
| P2_HD-R | YTGYTYTGCTTTTAAAGCATGG | |||
| 3 | P3_HD-F | GCTACTTTCAGACACTTAACAAATC | 491 | 552 |
| P3_HD-R | CCAAGAGAAATATGAACACATTTYC | |||
| 4 | P4_HD-F | TGCCATTCAATAyCGTCTTTG | 410 | 353 |
| P4_HD-R | AAATCCACAGTGATGAAATAACTTG | |||
| 5 | P5_HD-F | TCCTCCTCAATTTGCTCAATC | 217 | 262 |
| P5_HD-R | TGAATTTCCATTTGGTAATGAACT | |||
Figure 6 indicates the amplicons generated from P5_HD-F/ P5_HD-R primers for the 37 subjects. The figure indicates a single amplicon for each subject referring to ED (217 bp) or EH (262 bp) aligning with the results of the tRNA-linked STR approach (Figures 3 & 4), except for the three female subjects, e.g., 21, 27 and 35 where two amplicons referring to the two species were generated. The latter results indicates the coinfection with the two species in the three subjects. This conclusion was not reached using the tRNA-linked STR approach. We hypothesize that if other species-specific primers following the new approach were used, other species of Entamoaba might be found as a coinfection. The new approach has overcome the problem of primer mismatches and the occurrence of non-specific priming that could not be overcome utilizing the tRNA-linked STR approach despite our efforts to adjust the annealing temperatures. The major problem with the non-specific priming is the inability to recognize the right from the false products, thus, cannot refer a certain amplicon with a certain size to a certain species. The new approach is important in resolving this problem at the species level, while the tRNA-linked STR approach can still be a good choice for strain differentiation.
 |
Figure 6: Amplicons generated from P5_HD-F/ P5_HD-R primers for subjects infected with either Entamoeba species. Amplicons with 262 bp were generated from EH-infected subjects, |
Discussion
Strain differentiation of EH and ED are important in getting a better insights on the mysterious virulence of this parasite. Ali et al. [11] indicated that symptomatic and asymptomatic infections can be caused by genetically distinct strains. The STR-based strain differentiation might be a good approach in addressing unanswered questions on virulence of the parasite. Ali et al. [11] claimed that they were able to detect a unique feature of these two species and eliminated the potential problems caused by mixed infections or cultures. However, BLAST analysis for the first type of primers used in the present study indicates similar-sized amplicons of EH and ED. The latter conclusion indicates the possible confusion in characterizing the parasite in case of coinfection. Additionally, Feng et al. [16] detected seven different genotypes in ED from eight samples by sequence analysis of tRNA-linked short tandem repeats. These different genotypes were found within the same family. Therefore, we suggest that it is important first to use a more accurate species-specific markers way before studying strain differentiation and parasitology. Ali et al. [11] also claimed that they were able to distinguish clearly between the two species in fecal samples and eliminate the need for culturing the parasite using their approach. Our results indicated that the STR approach failed to clearly prove the existence of only one species in the studied samples as several amplicons were recovered for the different primers (either genus- or specific-primers) used with similar sizes in both species. Our results did not eliminate the possibility of coinfection in our samples.
There are several reports that address the high degree of STR length polymorphism among EH strains, even when the strains isolated from a restricted geographic location [13,17,18]. On top of this, our results indicated the existence of several amplicons that are common in subjects with diverse genetic makeup, therefore, we do not eliminate the possibility of having polymorphism in a given STR of the same parasite. The latter observations makes the use of strain differentiation alone more complicated and reject the claim of detecting patterns of transmission of this important disease and the epidemiological links between individual infection via the use of STR approach. Ghosh et al. [19] reported the occurrence of recombinational loss of a ribosomal DNA unit from the circular episome of Entamoeba histolytica HM-1:IMSS. Consequently, there is a possibility that recombinational gain can take place in circular episomes or in circular structures of tRNA as we claim. Investigations of EH genotypes in South Africa [18] and Vietnam [20] indicated similar STR patterns over the course of the same infection. These results are not controversy to ours as these patterns can still hold the possibility of having polymorphism not only among different STRs, but also within a given STR. This phenomenon is similar to the phenomenon of heteroplasmy in mitochondrial genomes. We hypothesize that if other species-specific primers following the new approach were used, other species of Entamoaba might be found as a coinfection. Finally, our results recommend the use of the new approach in species differentiation, while use tRNA-linked STR approach in strain differentiation.
Competing Interest
The authors declare that they have no competing interest.
Data Availability
Authors declare no unavailable data generated through or related to the course of this research.
Contributors
All authors have read and approved this manuscript. Conceived and designed the experiments: JS, SR, NH, AB. Performed the experiments: HY, NH, AA. Analyzed the data: HY, NH, AA, KA. Wrote the paper: JS, KA, NH, AB.
Funding
N/A
References
- Gathiram V, Jackson TFHG. A longitudinal study of asymptomatic carriers of pathogenic zymodemes of Entamoeba histolytica. S. Afr. Med. J. 1987;72:669–672.
- Lozano R, Naghavi M, Foreman K, et al. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the global burden of disease study 2010. Lancet 2012;380:2095-2128.
CrossRef - Liu J, Platts-Mills JA, Juma J, et al. Use of quantitative molecular diagnostic methods to identify causes of diarrhoea in children: a reanalysis of the GEMS case-control study. Lancet 2016;388:1291-1301.
CrossRef - Current status and progress: diarrhoea remains a leading killer of young children, despite the availability of a simple treatment solution. https://data.unicef.org/topic/child-health/diarrhoeal-disease/#. Updated March 2018.
- Swaminathan A, Torresi J, Schlagenhauf P, et al. GeoSentinel network. A global study of pathogens and host risk factors associated with infectious gastrointestinal disease in returned international travellers. J. Infect. 2009;59:19-27.
CrossRef - Shimokawa C, Kabir M, Taniuchi M, et al. Entamoeba moshkovskii is associated with diarrhea in infants and causes diarrhea and colitis in mice. J. Infect. Dis. 2012;206:744–751.
CrossRef - Ali IKM, Hossain MB, Roy S, et al. Entamoeba moshkovskii infections in children, Bangladesh. Emerg. Infect. Dis. 2003;9:580–584.
CrossRef - Ayeh-Kumi, PF, Ali IM, Lockhart LA, et al. Entamoeba histolytica: genetic diversity of clinical isolates from Bangladesh as demonstrated by polymorphisms in the serine-rich gene. Exp. Parasitol. 2001;99:80–88.
CrossRef - Shirley D-A, Farr L, Watanabe A review of the global burden, new diagnostics, and current therapeutics for amebiasis. Open Forum Infec. Dis. 2018;5:ofy161.
CrossRef - Oliveira FM, Neumann E, Gomes MA, et al. Entamoeba dispar: Could it be pathogenic. Parasitol. 2015;5:9-14.
CrossRef - Ali IKM, Zaki M, Clark CG. Use of PCR amplification of tRNA gene-linked short tandem repeats for genotyping Entamoeba histolytica. J. Clinical Microbiol. 2005;43:5842–5847.
CrossRef - Clark CG, Ali IKM, Zaki M, et al. Unique organization of tRNA genes in the protistan parasite Entamoeba histolytica. Mol. Biochem. Parasitol. 2006;146:24-29.
CrossRef - Haghighi A, Kobayashi S, Takeuchi T, et al. Remarkable genetic polymorphism among Entamoeba histolytica isolates from a limited geographic area. J. Clin. Microbiol. 2002;40:4081–4090.
CrossRef - Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: A laboratory manual, 2nd edition, Cold Spring Harbor, Laboratory Press, 1989.
- Kurtz S, Phillippy A, Delcher AL, et al. Versatile and open software for comparing large genomes, Genome Biol. 2004;5:R12.
CrossRef - Feng, M, Pandey K, Yanagi T, et al. Prevalence and genotypic diversity of Entamoeba species in inhabitants in Kathmandu, Nepal. Parasitol. Res. 2018;117:2467-2472.
CrossRef - Haghighi A, Kobayashi S, Takeuchi T, et al. Geographic diversity among genotypes of Entamoeba histolytica field isolates. J. Clin. Microbiol. 2003;41:3748–3756.
CrossRef - Zaki M, Reddy SG, Jackson TFHG, et al. Genotyping of Entamoeba species in South Africa: diversity, stability and transmission patterns within families. J. Infect. Dis. 2003;187:1860–1869.
CrossRef - Ghosh S, Zaki M, Clark CG, et al. Recombinational loss of a ribosomal DNA unit from the circular episome of Entamoeba histolytica HM-1:IMSS. Mol. Biochem. Parasitol. 2001;116:105–108.
CrossRef - Blessmann J, Ali IKM, Ton Nu PA, et al. Longitudinal study of intestinal Entamoeba histolytica infections in asymptomatic adult carriers. J. Clin. Microbiol. 2003;41:4745–4750.
CrossRef

This work is licensed under a Creative Commons Attribution 4.0 International License.





