Manuscript accepted on : 16 July 2016
Published online on: --
Plagiarism Check: Yes
Effects of Land use on the Structural Diversity of Soil Bacterial Communities in South Eastern Iran
Ghobad Jalali1, Amir Lakzian1,*, Alireza Astaraei1, Aliakbar Haddad-Mashadrizeh2, Mehdi Azadvar3 and Eisa Esfandiarpour4
1Department of Soil Science, Ferdowsi University of Mashhad.
2Department of Biology, Ferdowsi University of Mashhad.
3Department of Plant Protection, South Kerman Agricultural and Natural Resources Research and Education Center.
4Department of Soil Science, Vali-e-AsrUniversity of Rafsanjan.
Corresponding Author E-mail: alakzian@yahoo.com
DOI : http://dx.doi.org/10.13005/bbra/2325
ABSTRACT: Assessment of soil microbial community composition in arid and semi-arid regions is an important issue. The aim of this work was to analyze and compare the bacterial communities in three different neighboring land uses (orchard, farmand shrub land) in arid-region soils located in southeast of Iran using molecular biology-based. The analysis of the denaturing gradient gel electrophoresis(DGGE) of the polymerase chain reaction (PCR) amplified 16S rDNA fingerprints demonstrated that bacterial community composition in all land uses was different. Clustering analysis of the DGGE bands of 16S rDNAfragments revealed that bacteria in the three soil samples from each land use formed a separated cluster, indicating variation in species composition between land uses. However, the results showed that the species richness and Shannon-Wiener diversity index differed significantly among different land uses while Pielou's evenness index did not show any significant differences. These findings indicate that land use was one of the major factors in variation of species composition of soil bacterial communities.
KEYWORDS: Soil bacterial communities; Structural diversity; Land use; DGGE; Arid-region
Download this article as:| Copy the following to cite this article: Jalali G, Lakzian A, Astaraei A, Haddad-Mashadrizeh A, Azadvar M, Esfandiarpour E. Effects of Land use on the Structural Diversity of Soil Bacterial Communities in South Eastern Iran. Biosci Biotech Res Asia 2016;13(3). |
| Copy the following to cite this URL: Jalali G, Lakzian A, Astaraei A, Haddad-Mashadrizeh A, Azadvar M, Esfandiarpour E. Effects of Land use on the Structural Diversity of Soil Bacterial Communities in South Eastern Iran. Biosci Biotech Res Asia 2016;13(3). Available from: https://www.biotech-asia.org/?p=15727 |
Introduction
Soil microorganisms have avital role in numerous ecosystem processes such as soil organic matter decomposition, nutrient cycling, and organic carbon (OC) sequestration to play1-3.Bacteria are well known as the most abundant and diverse group of organisms living in soil4-6. Based on the majority of studies, physicochemicalcharacteristics of soil such as edaphic factors (soil type, texture, moisture, pH and nutrient availability) and land management practices are factors affecting the structural diversity of soil bacterial communities1-3,7-9.
There are a relatively large number of studies showing how changes in land use types can result in the alteration of soil bacterial communities and the biogeochemical processes they carry out. For example, Lauber et al. (2013)8 reported that there are various communities in soils of managed agricultural systems compared to unmanaged systems, with particular management practices, including liming and nitrogen fertilization, often having strongimpacts on the structure of microbial communities of soils.
However, most the relations between soil characteristics and microorganisms in just one region have been the focus of the majority of these studies. It has been shown that generalizing and transferring of findings obtained at one region with particular soil characteristics cannot be accurate for other regions with different soil characteristics10. So far, little is known about the existence of relationships between soil properties and biota common among different regions with their specific abiotic conditions3.
It is now well proved by microbiologists that just a small proportion of all bacteria have been isolated and characterized11. Comparing the percentage of culturable bacteria with total cell counts from various habitats revealed enormous differences12. New molecular techniques, targeting small subunit rRNA sequences by PCR amplification11, provide the most powerful tools for identification of bacterial diversity in environmental samples. Being based on the separation of polymerase chain reaction (PCR) amplicons of the same size but different sequences13, denaturing gradient gel electrophoresis (DGGE), introduced into microbial ecology by Muyzer et al. (1993)14, is probably the most frequently used technique among the culture-independent fingerprinting techniques12,15. As a well-established tool for studyingstructural diversity of microbial communities, many researchers have been using this technique in theirresearches thus far3,16-17.
For example, PCR-DGGE technique has been so far used for assessing the structural diversity among soil bacteria and fungi communities in rhizosphere which are the outcomes of differences in nutrient applications18. It has also been used forassessment of differences in soils under various agricultural management practices11,19-23 and soilsin different lands such asforest24-25,orchard16,pasture7,26 andcontaminatedlands15,27.
This research aimed to compare the structural diversity of soil bacterial communities in three land use types in southeastern Iran by using PCR-DGGE technique.
Material and methods
Study area
Jiroft plain with the elevation of 650 meters is one of the lowest plains in Iran. The plain with hot and dry climate is considered one of the most important agricultural plains of Kerman province. The study area located on geographical location 28˚ 28′ 40” to 28˚ 52′ 6” North and 57˚ 30′ 8” to 58˚ 4′ 27” East.
The county sees the average precipitation of 140 mm, average relative humidity of about 55% and the maximum and minimum temperatures of 48 and 1 degree Celsius, respectively. Soil thermal and moisture regimes of the region are hyperthermic and aridic, respectively.
The majority of the dominant cover of the pasture lands includes species of Haloxylon, Tamarisk, Jujube, Oleander and bitter melon. Furthermore, of the dominant agricultural and horticultural crops, onions, potatoes, wheat, barley, corn, citrus, palm and melons can be named.
Collection of soil samples
In Jiroft plain, a dominant geomorphologic unit (alluvial plain, located in the village of Ali Abad) containing shrubland that has not been cultivated so far because of water scarcity (Calligonumpersicum and etc.), field crops (at least in the last 50 years under fall planting of potatoes and onions, and in some years the spring planting of corn and etc.) and horticultural crops (more than 50 years under citrus cultivation) were selected.
The studied soils of the three neighboring land uses located on the alluvial plain were Aridisols. Three soil samples, each contained ten subsamples, were collected from A horizon (0-10 cm) across each land use on February 2015.
DNA extraction and PCR amplification
Total DNA from different soil samples was purified byusing the NucleoSpin®Soil kit (Macherey-Nagel, Duren, Germany) according to manufacturer’s protocols.
Targeting bacterial 16S rDNA was performed with the primer set of GC-341F(CGCCCGCCGCGCGCGGCGGGCGGGGCGGGGGCACGGGGGGCCTACGGGAGGCAGCAGCCTACGGGAGGCAGCAG) and 534R (ATTACCGCGGCTGCTGG) (A GC-rich clamp (40 bp) was added to the forward primerprevented the complete melting of the PCR amplicons during subsequent separation in DGGE)14.
Soil DNA was amplified in a Bio-Rad T100 thermal cycler(Bio-Rad Laboratories, CA,USA)with a PCR mixture (50 μl) containing 20 pmol of forwardand reverseprimers, 50 ng of DNA template, 25 μl of Taq DNA Polymerase 2X Master Mix Red (1.5mM MgCl2) (Ampliqon, Skovlunde, Denmark) and was filled up to the required volume with Milli-Q water.
The PCR cycles included initial denaturation at 94°C for 5 min, followed by a touchdown PCR protocolcontains: 20 cycles of 94ºC for 30S, 65-55°C(decreased 0.5°C per cycle) for 30S, 72ºC for 30S, and 15 cycles of 94ºC for 30S, 55°C for 30S, 72ºC for 30S and finally 72°C for 10 min. PCR products were checked for the expected size on 2% agarose gel with ethidium bromide under UV light.
DGGE
DGGE was performed using 8% polyacrylamide (37.5: 1, acrylamide:bisacrylamide) denaturing gel containing a 40-60% gradient(2.8 M urea/16% formamide to 4.2 M urea/24% formamide). About 25 μl of PCR products were applied on the denaturing gradient gel and the gel was then electrophoresed in 1X TAE buffer (40 mMTris base, 20 mM acetic acid, and 1 mM disodium EDTA, pH 8.3) at 60°C for 17 h at a constant voltage (100V) by using theDCodeTM Universal Mutation Detection System(Bio-Rad Laboratories, Hercules, CA, USA). After the electrophoresis, the gel was silver stained based on the protocol reported by Hwang et al. (2005)28.
Analysis of the DGGE profiles
GelCompar II 6.6 software was used for analysis of DGGE gel image. After removing of background intensity from lanes, the software performed a density profile through profiles, detected individual bands, and matched bands with the same position in different profiles.
Statistical procedures, except DGGE gel imaging, were carried out with IBM SPSS software. Means comparison, least significant differences (LSD) of 5% level, were calculated by a one-way ANOVA. Pairwise comparisons were performed using the Gabriel/Welsch’s multiple range test29. Three different ways were used for estimating of bacterial community compositionin the soil samples with GelCompar II 6.6 software: species richness (the number of bands present), species evenness or relative band intensity (Pielou’sevenness index) and species diversity (Shannon Wiener diversity index). Shannon- Wiener index was calculated according to the formula based on the relative band intensities as (H) = −ΣPilnPi = −Σ (Ni/N) ln (Ni/N). Pi was defined as Ni/N, where Ni is the area of a peak in intensity and N is the sum of all peak areas in the lane profiles11. Cluster analysis of the DGGE profiles was performed using the GelCompar II6.6 software. The similarity in the profiles of the bands was calculated on the basis of the Dice similarity coefficient with the UPGMA (Unweighted Pair-Group Method with Arithmetic Mean) clustering algorithm30.
Results
Physicochemical properties of soils
Three neighboring land use types were on the same edaphoclimatic conditions.
Farm and orchard lands had the same textural class of soil (loamy sand) while shrub land contained sandy soil. The mean valueof soil organic carbon was highest in orchard land (0.086%) and reduced to 0.043% and 0.030% in farm and shrub lands, respectively. The mean value ofpH in shrub land soils (8.17) was higher than those of farm (7.96) and orchard land soils (7.70). However, the highest and lowest percentage of CaCO3 in soil observed in farm (5.50%) and orchard lands (3.86%), respectively. In conclusion, results indicated the effects of land use on the selected physicochemical properties of soils.
Total DNA extraction
The total DNA was successfully purified from soil samples and was thus appropriate for the following PCR amplification. The results obtained from Nano drop showed that the means of total DNA yield were 32, 14 and 6.7 ng/ul in orchard, farm and shrub lands, respectively.
16S rRNA gene-based PCR products
As shown in figure 1, PCR amplification of V3 region of 16S rDNA fragments from the soil samples of three land uses produced a clear (∼230 bp) band against a 1kb ladder.
The product of this PCR amplicon was used for separation of different 16S rDNA nucleotide sequences by the DGGE technique.
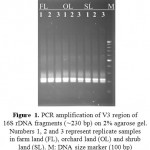 |
Figure 1: PCR amplification of V3 region of 16S rDNAfragments (∼230 bp) on 2% agarose gel.Numbers 1, 2 and 3 represent replicate samples in farm land (FL), orchard land (OL) and shrub land (SL). M:DNA size marker (100 bp) (DENAzist Asia Co. Iran). |
DGGE profiles of 16S rRNA gene-based PCR products
To assess the structural diversity of soil bacterial communities, DGGE banding patterns of V3 region of 16S rDNA amplified with primers GC341F and 534R were used.As shown clearly in Fig. 2, DGGE fingerprints of bacterial 16S rDNAamplicons from soils of the different land uses contained some common bands to all profiles while different bands observed with respect to relative band intensity.However, banding patterns among the replicate samples from the same land use were highly repetitive.A greater number of weaker, clear and unclear bands observed in the background showed the complex structure of bacterial communities in soil samples29.
Cluster analysis of the DGGE banding patterns based on unweighted pair-group method with arithmetic means (UPGMA)30indicated that the rates of similarity of the total genomic diversity among the nine soil samples collected from three land use types were 68.5%. Number and similarity of bands observed in each profile with respect to other profiles is the basis of this analysis.Therefore,in order to evaluate the structure of the bacterial communities among the soil samples, the obtained clusters which are the indices of similarity or dissimilarity16 were used.
In general, results indicated that dominant bacterial communities had different structures and the three soil samples from every land use formed a separate cluster (fig. 2). The cluster I consists of 16S rDNA bands derived from the orchard land that had the minimum similarity index of 74.04%, while the three soil samples of farm and shrub lands formed the clusters II and III, respectively. Soil samples from farm land had greater similarity with each other (shared at least 80.09% similarity) compared to those from the shrub land (76.89% similarity).
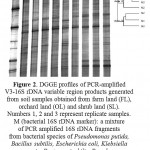 |
Figure 2: DGGE profiles of PCR-amplified V3-16S rDNAvariableregion products generated from soil samples obtained from farm land (FL), orchard land (OL) and shrub land (SL). Numbers 1, 2 and 3 represent replicate samples.M (bacterial 16S rDNA marker): a mixture of PCR amplified 16S rDNA fragments from bacterial species ofPseudomonas putida, Bacillus subtilis, Escherichia coli, Klebsiella pneumonia, Proteus mirabilis, Pseudomonasaeruginosa, Acinetobacterbaumannii, and Salmonella entericaserovarTyphi. Comparative DGGE analysis were made by using Dice similarity coefficient based on UPGAMA cluster analysis (n = 3) |
The statistical analysis ofdiversity indices
For evaluatingthe significant differences in diversity indices resulted from DGGE banding patterns, one-way ANOVA was used.The results of analysis showed that the values of species richness (the number of bands present) and Shannon–Wiener diversity index were significantly (p≤0.05) different among different types of land use. Pairwise comparisons indicated that the mean of species richness in the profiles of the shrub land (33.33) was significantly lower than those of farm (45.33) and orchard (39.67) lands. However, the mean of Shannon–Wiener index in the profiles of the shrub land (1.35) was significantly lower than those in the farm (1.55) and orchard (1.49) profiles. However, compared to the orchard, the farm profiles showed higher, but not significant difference.Statistical analysis of species evenness (Pielou’sevenness index) showed that land use did not have significant effect on bacterial evenness (table 1).
Table 1: The meansof calculated diversity indices (Shannon–Wiener diversity index, species richness and Pielou’s evenness index) from DGGE banding patterns
| Land use | Shannon–Wiener diversity index | Species richness | Pielou’sevenness index |
| Orchard land (OL) | 1.49a | 39.67a | 0.927a |
| Farm land (FL) | 1.55a | 45.33b | 0.937a |
| Shrub land (SL) | 1.35b | 33.33c | 0.909a |
Discussion
Because of the complex matrix and physiochemical characteristics, soils are believed to be among the most difficult environments to work31.These specific properties and surplus methodological challenges create several critical topics for soil metagenomics studies that may misrepresent the real microbial diversity from such soils, and therefore, influence our understanding about the structure and function of soil microbial communities32. Soil sampling is the first issue to be considered33. In our study, the three neighboring land uses located in one geomorphologic unit (alluvial plain) were selected in order to show that the discrepancies in soil properties are land use- based differences.
The DNA band pattern acquired by DGGE can be used as a semi-quantitative evaluation of bacterial diversity19; therefore it was used to evaluate the diversity of soil bacterial community under various land use types in arid region of the Jiroft County, southeastern Iran.
The different banding patterns observed in the soil bacterial DNA profiles, with many low-intensity and closely-spaced bands interspersed with a few bright dominant bands, were probably the result of high heterogeneity existed in extracted DNA, showing the complexity of the bacterial community composition22.
The similarity observed in the band profiles of the replicate samples from the same land use indicated that there were greater differences regarding the structure of bacterial population between the different land uses than within the same one29. Furthermore, there were clear structural shifts between agricultural (farm and orchard) and shrub land soils. Under the conditions of using directly isolated soil DNA,which was the same as our research, Crecchio et al. (2004)34 reported the same type of banding pattern on responses of soil microbial community to different management practices. They suggested that the presence of bright bands perhaps indicated that a limited number of dominant and ecologically well-adapted bacterial types existed in the soil. On the other hand, the numerous light bands possibly showed that abundant populations characterized each soil34.
Based on the results, these soils contained highly complex fingerprints, and thusaccurate calculation of the total number of bands was rather impossible. DGGE fingerprinting is known to determine only the predominant members in a community so it does not show the full complexity of the system35. In fact, community fingerprint data cannot provide an accurate estimate for evaluation of diversity indices in complex communities14. Therefore, the bands obtained from a DGGE gel in this study were considered as the dominating bacterial members in the soils of the studied area1.
We in cluster analysis observed a separate cluster for each land use concludingthat land use type could have significant effect on the soil bacterial community.
This could be due to many factors interacting with each other, for example, the soils being covered with different types of plants36-37. Other difference between the three land uses might be related to the quality and quantity of rhizodepositions16.
The difference in bacterial communities between cultivated lands (orchard and farm) and shrub land might be partly the result of land management applied in cultivated lands37. The idea that soil management practices and several environmental factors affect the structure of soil microbial community hasbeen reported by many researchers.For example, a studyconducted byBossio et al. (2005)20 showed that DGGE analysis of amplified 16S rDNA fragments revealed different structures in microbial community based on different soil management practices. Moreover, Marschner et al. (2001)36 studied the effects of five different treatments on soil bacterial and eukaryotic community structures and showed that different treatments caused great changes in both structures, especially in soil containing low organic C content.Additionally, the pH of soil in cultivated lands was lower than that of studied shrub land, whichcould be attributed to the long-term application of chemical fertilizers causing a decline in the pH of soil38-39. As several researches stated, the structures of soil microbial community were largely determined by soil pH40-41. Therefore, soil pH could be another driving factor changing structures of bacterial community between studied shrub land and cultivated lands.
According to our results,there was significant difference in Shannon-Wiener diversity index and species richness (the number of bands present) but not in species evenness (Pielou’s evenness index) in soil bacterial communities across three land use types.Based on the Shannon-Wiener index, it was found that farm and orchard lands reflected greater diversity compared to shrub lands. Pairwise comparisons separated shrub land bacterial communities from those of orchard and farm lands, although no difference was found in pairwise comparisons of the communities between orchard and farm lands. Because of a relatively low average annual precipitation (140 mm) in arid-region of Jiroft County, cultivated lands (citrus orchard and field crops) indicated the greater influences on soil bacterial communities compared to shrub land.Our results are close to the results reported by Koberl et al. (2011)42in arid soils of Egypt, where theyconcluded that the diversity of bacterial communities in agricultural land was higher than in desert land.
Several studies have reported that various land uses changed soil microbial diversity. For example, comparing the effects of long-term usage of manure and chemical fertilizers on soil bacterial community structure, Sun et al. (2004)43 found that the number of DGGE band in NPK-treated soil was significantly lower than that in manure-treated soil and in conclusion claimed that bacterial community structure diversity revealed a significant decrease by application of chemical fertilizers. Moreover, Waid (1999)44claimed that vegetation type,quantity and chemical compositions might be really effective in determining the diversity of soil microbial community.
In overall, most of the researchers expressed that vegetation type is an important factor in determining soil microbial community diversity36,45-48.
Conclusion
The structural diversity of soil bacteria communities in thestudied arid-regionof southeastern Iranhas been affected by a combination of long-term land use and soil physicochemicalcharacteristics.Based on our findings there were significant differences in the number of bands observed on DGGE and diversity index among three neighboring land use types, so that each land use had different predominant bacteria.
As a relatively simple and cost-effective method for determining structural diversity of soil bacterial communities, PCR-DGGE was used successfully for classification of multiple soil samples in the three land use types.
This study reports, for the first time, a comparative analysis of the changes in the soil bacterial diversity associated with land use types. The current study can provide a starting point for further investigations leading to a better understanding of the effect of land use changes on soil microbial communities in arid regions of the Iran.
Acknowledgments
The project (3/31214) was financially supported by Ferdowsi University of Mashhad, Iran.
References
- Ye, R., Wright, A.L., Inglett, K., Wang, Y., Ogram, A.V., Reddy, K.R. Land use effects on soil nutrient cycling and microbial community dynamics in the everglades agricultural area, Florida. Commun. Soil Sci. Plan, 2009; 40: 2725-42.
CrossRef - Hu, J., Lin, X., Wang, J., Dai, J., Chen, R., Zhang, J., Wong, M.H. Microbial functional diversity, metabolic quotient, and invertase activity of a sandy loam soil as affected by long-term application of organic amendment and mineral fertilizer. J. Soil Sediment, 2011; 11: 271-8.
CrossRef - Herold, N., Schoning, I., Gutknecht, J., Alt, F., Boch, S., Muller, J., Oelmann, Y. Socher, S.A., Wilcke, W., Wubet, T., Schrumpf, M. Soil property and management effects on grassland microbial communities across a latitudinal gradient in Germany. Appl. Soil Ecol., 2014; 73: 41-50.
CrossRef - Martınez, V.A., Dowd, S., Sun, Y., Allen, V. Tag-encoded pyrosequencing analysis of bacterial diversity in a single soil type as affected by management and land use. Soil Biol. Biochem., 2008; 40: 2762-70.
CrossRef - Moorthy, K. Isolation of Soil Bacteria for Bioremediation of Hydrocarbon Contamination. Biosci.Biotechnol. Res. Asia, 2010; 7(2).
- Eswar Ganesh Babu, T., Mastan, S.A. Isolation and Characterization of Bio-Degrading Bacteria from Soil Samples. Biosci.Biotechnol. Res. Asia, 2011; 8(1).
- Lauber, C.L., Strickland, M.S., Bradford, M.A., Fierer, N. The influence of soil properties on the structure of bacterial and fungal communities across land-use types. Soil Biol. Biochem., 2008; 40: 2407-15.
CrossRef - Lauber, C.L., Ramirez, K.S., Aanderud, Z., Lennon, J., Fierer, N. Temporal variability in soil microbial communities across land-use types. ISME J., 2013; 7: 1641-50.
CrossRef - Ramirez, K.S., Lauber, C.L., Knight, R., Bradford, M.A., Fierer, N. Consistent effects of nitrogen fertilization on soil bacterial communities in contrasting systems. Ecology, 2010; 91: 3463-70.
CrossRef - Gianfreda, L., Rao, M.A., Piotrowska, A., Palumbo, G., Colombo, C. Soil enzyme activities as affected by anthropogenic alterations: intensive agricultural practices and organic pollution. Sci. Total Environ., 2005; 341: 265-79.
CrossRef - Tao, S.Q., Xia, Q., Zhu, L., Chen, J.J., Wang, Y.N., Qin, B. Analysis of the bacterial communities in lime concretion black soil upon the incorporation of crop residues. Open J. Soil Sci., 2012; 9: 312-19.
CrossRef - Muyzer, G., Smalla, K. Application of denaturing gradient gel electrophoresis (DGGE) and temperature gradient gel electrophoresis (TGGE) in microbial ecology. Anton. Leeuw. INT. J. G., 1998; 73: 127-41.
- Valaskova, V., Baldrian, P. Denaturing gradient gel electrophoresis as a fingerprinting method for the analysis of soil microbial communities. Plant Soil Environ., 2009; 55(10): 413–23.
CrossRef - Muyzer, G., de Waal, E.C., Uitterlinden, A.G. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rDNA. Appl. Environ. Microb., 1993; 59: 695-700.
- Li, Z., Xu, J., Tang, C., Wu, J., Muhammad, A., Wang, H. Application of 16S rDNA-PCR amplification and DGGE fingerprinting for detection of shift in microbial community diversity in Cu-, Zn-, and Cd-contaminated paddy soils. Chemosphere, 2006; 62: 1374-80.
CrossRef - Xue, D., Yao, H., Huang, C. Microbial biomass, N mineralization and nitrification, enzyme activities, and microbial community diversity in tea orchard soils. Plant Soil, 2006; 288: 319-31.
CrossRef - Sharma, S.K., Ramesh, A., Sharma, M.P., Joshi, O.P., Govaerts, B., Steenwerth, K.L., Karlen, D.L.: Microbial community structure and diversity as indicators for evaluating soil quality. In: Biodiversity, Biofuels, Agroforestry and Conservation Agriculture, Sustainable Agriculture Reviews 5 (Lichtfouse E., ed). Springer Science + Business Media B.V., 2011; pp. 317-358.
- Smalla, K., Wieland, G., Buchner, A., Zock, A., Parzy, J., Kaiser, S., Roskot, N., Heuer, H., Berg, G. Bulk and rhizosphere soil bacterial communities studied by denaturing gradient gelelectrophoresis: plant-dependent enrichment and seasonal shifts revealed. Appl. Environ. Microb., 2001; 67: 4742-51.
CrossRef - Dilly, O., Bloem, J., Vos, A., Munch, J.C. Bacterial diversity in agricultural soils during litter decomposition. Appl. Environ. Microb., 2004; 70: 468-74.
CrossRef - Bossio, D.A., Girvan, M.S., Verchot, L., Bullimore, J., Borelli, T., Albrecht, A., Scow, K.M., Ball, A.S., Pretty, J.N., Osborn, A.M. Soil microbial community response to land use change in an agricultural landscape of Western Kenya. Microb. Ecol., 2005; 49: 50-62.
CrossRef - Kamaa, M., Mburu, H., Blanchart, E., Chibole, L., Chotte, J.L., Kibunja, C., Lesueur, D. Effects of organic and inorganic fertilization on soil bacterial and fungal microbial diversity in the Kabete long term trial, Kenya. Biol. Fert. Soils, 2011; 47: 315-21.
CrossRef - Ding, G.C., Piceno, Y.M., Heuer, H., Weinert, N., Dohrmann, A.B., Carrillo, A., Andersen, G.L., Castellanos, T., Tebbe, C.C., Smalla, K. Changes of soil bacterial diversity as a consequence of agricultural land use in a semi-arid ecosystem. PLoS ONE, 2013; 8(3): e59497.
- Bevivino, A., Paganin, P., Bacci, G., Florio, A., Pellicer, M.S., Papaleo, M.C., Mengoni, A., Ledda, L., Fani, R., Benedetti, A., Dalmastri, C. Soil bacterial community response to differences in agricultural management along with seasonal changes in a Mediterranean region., PLoS ONE, 2014; 9(8): e105515.
- Dong, X., Ying, Y.H., Yong, G.E., Yong, H.C. Soil microbial community structure in diverse land use systems: A comparative study using Biolog, DGGE, and PLFA analyses. Pedosphere, 2008; 18(5): 653-63.
CrossRef - Singh, S.S., Schloter, M., Tiwari, S.C., Dkhar, M.S. Diversity of community soil DNA and bacteria in degraded and undegraded tropical forest soils of North-Eastern India as measured by ERIC–PCR Fingerprints and 16S rDNA-DGGE Profiles. J. Biol. Environ. Sci., 2011; 5(15): 183-94.
- Ritz, K., Mac Nicol, J.W., Nunan, N., Grayston, S., Millard, P., Atkinson, A., Gollotte, A., Habeshaw, D., Boag, B., Clegg, C.D. Spatial structure in soil chemical and microbiological properties in upland grassland. FemsMicrobiol. Ecol., 2004; 49:191-205.
CrossRef - Cycon, M., Markowicz, A., Seget, Z.P. Structural and functional diversity of bacterial community in soil treated with the herbicide napropamide estimated by the DGGE, CLPP and r/K-strategy approaches. Appl. Soil Ecol., 2013; 72: 242-50.
CrossRef - Hwang, S.Y., Jin, L.T., Yoo, G.S., Choi, J.K. Silver staining method for DNA in polyacrylamide gels using eriochrome black T as a silver ion sensitizer. Electrophoresis, 2006; 27: 1744-48.
CrossRef - Wallis, P.D., Haynes, R.J., Hunter, C.H., Morris, C.D. Effect of land use and management on soil bacterial biodiversity as measured by PCR-DGGE. Appl. Soil Ecol., 2010; 46: 147-50.
CrossRef - Montecchia, M.S., Correa, O.S., Soria, M.A., Frey, S.D., Garcia, A.F., Garland, J.L. Multivariate approach to characterizing soil microbial communities in pristine and agricultural sites in Northwest Argentina. Appl. Soil Ecol., 2011; 47: 176-83.
CrossRef - Delmont, T.O., Robe, P., Cecillon, S., Clark, I.M., Constancias, F., Simonet, P., Hirsch, P.R., Vogel, T.M. Accessing the soil metagenome for studies of microbial diversity. Appl. Environ. Microb., 2011; 77: 1315-24.
CrossRef - Lombard, N., Prestat, E., van Elsas, J.D., Simonet, P. Soil specific limitations for access and analysis of soil microbial communities by metagenomics. FemsMicrobiol. Ecol., 2011; 78:31-49.
CrossRef - Rampelotto, P.H., Ferreira, A.D.S., Barboza, A.D.M., Roesch, L.F.W. Changes in diversity, abundance, and structure of soil bacterial communities in Brazilian Savanna under different land use systems. Microb. Ecol., 2013; 66(3): 593-607.
CrossRef - Crecchio, C., Curci, M., Pizzigallo, M.D.R., Ricciuti, P., Ruggiero, P. Effects of municipal solid waste compost amendments on soil enzyme activities and bacterial genetic diversity. Soil Biol. Biochem., 2004; 36: 1595-1605.
CrossRef - Ascher, J., Ceccherini, M.T., Chronakova, A., Jirout, J., Borgogni, F., Elhottova, D., imek, M.S., Pietramellara, G. Evaluation of the denaturing gradient gel electrophoresis apparatus as a parameter influencing soil microbial community fingerprinting. World J.Microb. Biot., 2010; 26(9): 1721-26.
CrossRef - Marschner, P., Yang, C.H., Lieberei, R., Crowley, D.E. Soil and plant specific effects on bacterial community composition in the rhizosphere. Soil Biol. Biochem., 2001; 33: 1437-45.
CrossRef - Miethling, R., Ahrends, K., Tebbe, C.C. Structural differences in the rhizosphere communities of legumes are not equally reflected in community level physiological profiles. Soil Biol. Biochem., 2003; 35: 1405-10.
CrossRef - Liu, E., Yan, C., Mei, X., He, W., Bing, S.H., Ding, L., Liu, Q., Liu, S., Fan, T. Long term effect of chemical fertilizer, straw, and manure on soil chemical and biological properties in northwest China, Geoderma, 2010; 158: 173-80.
CrossRef - Shen, J.P., Zhang, L.M., Guo, J.F., Ray, J.L., He, J.Z. Impact of long-term fertilization practices on the abundance and composition of soil bacterial communities in Northeast China, Appl. Soil Ecol., 2010; 46: 119-24.
CrossRef - Fierer, N., Jackson, R.B. The diversity and biogeography of soil bacterial communities. P. Natl. Acad. Sci. U.S.A., 2006; 103: 626-31.
CrossRef - Wakelin, S.A., Macdonald, L.M., Rogers, S.L., Gregg, A.L., Bolger, T.P., Baldock, J.A. Habitat selective factors influencing the structural composition and functional capacity of microbial communities in agricultural soils, Soil Biol. Biochem., 2008; 40: 803-13.
CrossRef - Koberl, M., Muller, H., Ramadan, E.M., Berg, G. Desert farming benefits from microbial potential in arid soils and promotes diversity and plant health. PLOS ONE, 2011; 6(9): e24452.
CrossRef - Sun, H.Y., Deng, S.P., Raun, W.R. Bacterial community structure and diversity in a century-old manure-treated agroecosystem. Appl. Environ. Microb., 2004; 70: 5868-74.
CrossRef - Waid, J.S. Does soil biodiversity depend upon metabiotic activity and influences? Appl. Soil Ecol., 1999; 13:151-8.
CrossRef - Grayston, S.J., Griffith, G.S., Mawdsley, J.L., Campbell, C.D., Bardgett, R.D. Accounting for variability in soil microbial communities of temperate upland grassland ecosystems. Soil Biol. Biochem., 2001; 33: 533-51.
CrossRef - Berg, G., Roskot, N., Steidle, A., Eberl, L., Zock, A., Smalla, K. Plant dependent genotypic and phenotypic diversity of antagonistic rhizobacteria isolated from different Verticillium host plants. Appl. Environ. Microb., 2002; 68: 3328–38.
CrossRef - Guanghua, W., Junjie, L., Xiaoning, Q., Jian, J., Yang, W., Xiaobing, L. Effects of fertilization on bacterial community structure and function in a black soil of Dehui region estimated by Biolog and PCR-DGGE methods., Acta Ecol. Sin., 2008; 28(1): 220-6.
CrossRef - Yu, Z., Wang, G., Jin, J., Liu, J., Liu, X. Soil microbial communities are affected more by land use than seasonal variation in restored grassland and cultivated Mollisols in Northeast China. Eur. J. Soil Biol., 2011; 47: 357-63.
CrossRef

This work is licensed under a Creative Commons Attribution 4.0 International License.





