Manuscript accepted on : 04 February 2016
Published online on: 25-03-2016
Detection of Escherichia coli Pathotypes from the Cases of Diarrhea
Maryam Zarringhalam1, Hossein Goudarzi2*, Mohammad Reza Nahaei3, Mojgan Bandehpour4 and Gholamhassan Shahbazi3
1Department of Microbiology, International Branch of Shahid Beheshti University of Medical Sciences, Tehran, Iran. 2Department of Microbiology, Shahid Beheshti University of Medical Sciences, Tehran, Iran. 3Department of Microbiology, Tabriz University of Medical Sciences, Tabriz, Iran. 4Department of Biotechnology, Shahid Beheshti University of Medical Sciences, Tehran, Iran. Corresponding Authro Email: medicalopto@yahoo.com
DOI : http://dx.doi.org/10.13005/bbra/2028
ABSTRACT: Escherichia coli is a member of the normal microbiota,and also a common inhabitant of the human and warm-blooded animal intestinal tract, which several pathogenictypes of it can cause different diseases.One hundred and fourty seven E. coli isolates were obtained from the cases of diarrhea during 2013-2014. In order to detect E. coli pathotypes a mixture of nine primer pairs was used in three multiplex PCR assays. The PCR assays detected 26 ETEC, 20 EAEC, 11 EHEC, 9 EPEC and 2 EIEC. All E. coli strains were serogrouped by a latex agglutination test using a commercial antiserum. According to the results, 53 isolates were positive and according to kit regents, 3 different groups were identified. The findings also indicated that the E. coli strains recovered in this study expressed high levels of resistance to antimicrobials that are commonly used in clinical medicine. In conclusion,differentiation between the diarrheagenic E. coli pathotypes is very importantsince they are involved in cancer or cellular senescence. It was concluded that at attempts at economy are better conducted via finding appropriate criteria for requesting a stool culture rather than at limitation of the microbiological evaluation of specimens that are subjected.
KEYWORDS: Serogrouping; Multiplex; PCR; Detection; Escherichia coli
Download this article as:| Copy the following to cite this article: Zarringhalam M, Goudarzi H, Nahaei M. R, Bandehpour M, Shahbazi G. Detection of Escherichia coli Pathotypes from the Cases of Diarrhea. Biosci Biotech Res Asia 2016;13(1) |
| Copy the following to cite this URL: Zarringhalam M, Goudarzi H, Nahaei M. R, Bandehpour M, Shahbazi G. Detection of Escherichia coli Pathotypes from the Cases of Diarrhea. Biosci Biotech Res Asia 2016;13(1). Available from: https://www.biotech-asia.org/?p=7430 |
Introduction
Escherichia coli are commensal bacteria of the human gut microbiota, but some pathogenic strains have acquired the ability to induce chronic inflammation and/or produce toxins, such as cyclomodulin, which could participate in the carcinogenesis process. It was explained that the E. coli population associate with mucosa of patients with colon cancer in relation to clinicopathologic characteristics1.
The widespread species Escherichia coliare categorized into six pathotypes, which are associated with diarrhea and collectively are referred to as diarrheagenic E. coli: enterotoxigenic E. coli (ETEC), which produces one or more enterotoxins that are heat labile LT (lt) or heat stable ST (st); enteropathogenic E.coli (EPEC), which has a pathogenicity island that encodes a series of proteins involved in the attaching and effacement lesions of the intestinal microvilli of the host cell; and the presence of the large EPEC adherence factor (EAF) plasmid (eae), on which also the cluster of genes encoding bundle-forming pilli (bfp) is present. Based on these, EPEC strains are classified as typical when they possess the EAF plasmid, whereas atypical EPEC strains don’t possess the EAF plasmid; enterohemorrhagic (EHEC) or Shiga toxin producing E.coli (STEC) is characterized by the production of two strong cytotoxins denominated Shiga-like toxins 1 and 2 (stx1 and stx2).Like EPEC, EHEC elicits an A/E lesion of the intestinal mucosa, a phenotype that requires a functional eaeA chromosomal gene2, 3.
The three other categories seem to be less prevalent. Enteroinvasive E. coli (EIEC) has biochemical, physiological, and genetic properties similar to those of Shigella, invading the epithelial cells of the colon, where it proliferates and causes necrosis of the tissue. Invasion plasmid antigen H (ipaH) found in EIEC and Shigella;3enteroaggragative E.coli (EAEC) strainsare characterized byan aggregative adherence (AA) pattern on cultured epithelial cell and produce fimbrial colonization factors called aggregative adherence factors (AAF)4.The most recently characterized category corresponds to diffusely adherentE.coli (DAEC), strains that are capable of adhering to HEP-2 cells in a nonlocalized pattern.The “gold standard” method for detection of DAEC strains is based on the diffusely adherent pattern in the HEp-2 adherence assay or by detection of the gene daaE that is necessary for the expression of the F1845 fimbriae2.
These E. colipathotypes are commonly isolated from patients with gastroenteritis in the developing world. However, they are not routinely sought as stool pathogens in clinical laboratories. Some of these pathogens respond to antimicrobial agents, while for others (e.g., Shiga toxin-producing E. coli [STEC]), antibiotics should be avoided. Since the time frame in which treatment choices must be made is short, there is a need for a rapid, sensitive, and inexpensive detection technique5.
Infections caused by diarrheagenic E.coli (DEC)strains are important;therefore, it is important to detect them at the onset and pursue necessary preventive measures. For many years, laboratory detection of food–borne pathogens has relied on direct isolation, which is still the preferred method, as it represents the gold standard as compared to newly developed rapid detection methods.However, several methods available for rapid detection are discussed in order to shorten the detection time and begin therapeutic measures6, 7.
Among all diagnostic techniques such as culture, serology and molecular methods, the latter has high sensitivity, specificity and safety. Nonetheless, culture and serology methods remain popular because of their simplicity. Serotyping and biochemistry have been widely applied in the diagnosis of gastrointestinal pathogens, but they cannot be used for definite detection of DEC groups. Therefore, identification of the characteristic virulence genes is a better choice for DEC diagnostics6, 8.
Little is known about the prevalence of E.colipathotypes in Iran. It has been estimated that diarrheagenicE. coli are among the most prevalent causative agents in acute diarrhea9, 10.However, there are no well-documented mortality figures for E. coli-induced disease, because the microbiologic diagnosis cannot be done easily in many settings. E. coli-related deaths at present would be counted as diarrheal and colon cancer deaths in many countries1. So, knowledge of the status of the E.coli pathotypes in Iran is important for planning appropriate public health programs for controlling the disease. Therefore, the aim of this study was to evaluate the usefulness of serology and PCR methods in detecting the E.coli strains isolated in Iran.
Materials and Methods
Clinical specimens
One hundred and fourty seven E.coli isolates were obtained from the cases of diarrhea in the hospitals of Tabriz (Iran)within 2013-2014. These isolates were identified by conventional methods and stored at -70°C in TSB (Merck, Germany) restraining 20% glycerol until required12.
Bacterial Strains
Diarrheagenic E. coli reference strains 933 J (stx1, stx2, eae), 2348/69 (eae,bfp), H10407 (st, lt), O42 (ipaH), and F-1845 (daaE) were used as positive controls. One hundred andforty sevenE. coli strains were analyzed, including strains representing all of the currently recognized six classes of diarrheagenic E. coli as well as the commensal organisms. The prototypical strains currently used in the laboratories worldwide include enterotoxigenic E. coli (ETEC) H10407; enteropathogenic E. coli (EPEC) 2348/69; enteroaggregative E. coli (EAEC) O42; E. coli (EIEC) EI-34 andE. coli (DAEC) F-1845 (daaE).Reference strains were provided bythe Center for Vaccine Development (CVD), University of Maryland; MP, Microbiology Program, University of Chile.
Extraction of DNA
A sweep of about five E.coli like colonies was cultured in Tripticase Soy Broth (TSB) for 18h. The DNA was extracted using Promega genomic DNA extraction kit according to the manufacture’s protocol (A11125, USA).
PCR primers Design
PCR primers were designed based on the virulence gene sequences resumed from the National Center for Biotechnology Information (NCBI) database2. The obtained amplicons ranged from 100 to 900 bp in length. All primers were obtained from GeNetBio Inc., Korea (Table1).
Multiplex PCR Assays
The multiplex PCR assays were carried out using a 2X PCR Master Mix, which is an optimized premixed with 2X concentrated solutions of Taq DNA polymerase, reaction buffer, MgCl2 and dNTPs. The 2X PCR Master Mix contains all components for PCR, except DNA template and primers. The DNA templates were subjected to multiplex PCR with specific primers (Table2).Three multiplex PCRs were standardized for identification of the DECs.
Multiplex PCR Assay 1
The reaction mixture of optimized protocol was carried out with a 25 μl mixture of2X PCR Master Mix containing: 5 μl of the DNA template and 1 μM of each of the primers (GeNetBio Inc., Korea), i.e. lt and st for ETEC isolates, and ipaHfor EIEC isolates. The cycling conditions in Bio Rad T100TM thermal cycler were as follows: 95°C for 5 min for one cycle followed by 35 cycles of 95°C for 45sec, 49°C for 45sec, and72°C for 1 min and 72°C for 10 min.
Multiplex PCR assay 2
The protocol used was as explained above for multiplex PCR 1except that the primers were eae and bfp for EPEC isolates, and eae, stx1 and stx2 for EHEC isolates. The thermocycling conditions were as follows: 95°C for 5 min for one cycle followed by 38 cycles of 95°C for 1 min, 53°C for 1 min, 72°C for 1min and 72°C for 10 min.
Multiplex PCR Assay 3
The multiplex PCR assay 3 was performed with the daaE and aggR (transcriptional activator for the AAFs) primers for DAEC and EAEC. The amplification conditions were as follows: an initial denaturation step at 95°Cfor 5 min was followed by 39 cycles of 95°C for 45 sec, annealing at 55°C for 45 sec and elongation at 72°C for 1 min. Afinal elongation step was executed at 72°C for 10 min. PCR products were evaluated on a 1.5 % (w/v) agarose gel (Ultrapure Agarose).The DNA bands were visualized under UV light after staining the gel with ethidium bromide (Fermentas Inc., Maryland, USA)3, 13.
Serotyping
Serotyping is based on the use of specific antisera and the detection of somatic O and Kantigens expressed by E.colipathotypes (Table1).O-K polyclonal antisera (Sifin Kit) were used for slide agglutination with live cultures, and the reaction was observed after 5-10 sec14.
Antibiotic susceptibilitytests
Susceptibility testing was conducted by Kirby-Bauer disk diffusionaccording to the guidelines of the Clinical and Laboratory Standards Institute (CLSI). Susceptibility of E.coli isolates was tested to the commonly used antibiotics such as Cefoxitin, Ceftazidime, Cefotaxime, Cefepime, Gentamicin, Kanamycin, Co-trimoxazole, Ampicillin, Imipenem, and Ciprofloxacin (Table 3). E. coli ATCC 25922 was used as quality control11, 15.
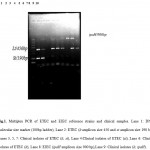 |
Figure 1: Multiplex PCR of ETEC and EIEC reference strains and clinical samples. Lane 1: DNA molecular size marker (100bp ladder), Lane 2: ETEC (lt amplicon size 450 and st amplicon size 190 bp), Lanes 3, 5, 7: Clinical isolates of ETEC (lt, st), Lane 4:Clinical isolates of ETEC (st), Lane 6: Clinical isolates of ETEC (lt), Lane 8: EIEC (ipaH amplicon size 900 bp),Lane 9: Clinical isolates (lt, ipaH).
|
 |
Figure 2: Multiplex PCR of EHEC and EPEC reference strains and clinical samples. Lanes 1, 9: Negative control, Lanes 2, 3: Clinical isolates of EHEC (stx1,stx2), Lanes 4, 6: EHEC (stx1, eae,stx2), Lane 5: DNA molecular size marker (100bp ladder), Lane 7: EPEC (eae, bfp amplicon size 326 bp), Lane 8: Clinical isolates of EHEC (stx1amplicon size 388, eaeamplicon size 570 and stx2 amplicon size 807bp), Lane10: aEPEC (eae), Lane 11: Clinical isolates of aEPEC (eae).
|
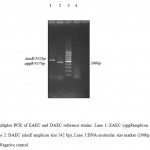 |
Figure 3: Multiplex PCR of EAEC and DAEC reference strains. Lane 1: EAEC (aggRamplicon size 457 bp), Lane 2: DAEC (daaE amplicon size 542 bp), Lane 3:DNA molecular size marker (100bp ladder), Lane 4: Negative control.
|
Results
All E.coli strains were serogrouped by a latex agglutination test using a commercial antiserum. As demonstrated in Table 1, of 147E.coli isolates subjected to latex agglutination test, 53 isolates were positive, and according to the kit regents, three different groups were identified;13 (8.84%) strains showed positive reaction with antiserum 1, and in 10(6.80%) strains, agglutination with antiserum 2was observed. Also, in 30(20.4%) strains, positive response was identified in antiserum 3. It was detected that 6 ETEC, 4 EPEC, 5 EHEC, 5 EAEC and 1 EIEC were not typeable with O-K polyclonal antisera. As evident in Tables 1 and 4, the most commonly detected E.coli seroroups were O25, O78, O103, O118, O124, O145, O157 and O164, while 23 strains were untypeable by the available antisera. As a result, thehighest percentage of ETEC, EPEC and EHEC isolates showed positive response with antiserum 3. One of the E.coli isolates showed positive reaction with all of the kit reagents, but no virulence factor genes were isolated in the PCR assays. Also seven isolates were positive in the latex agglutination test; however, virulence genes were not detected with our primers.
To demonstrate the utility of multiplex PCR assays, 147 E.coli strains isolated from diarrheic patients were subjected to the multiplex PCR, and the results were compared with those obtained by single PCRs (Table 4). The PCR assays detected 26 ETEC, 20 EAEC, 11 EHEC, 9 EPEC, and two EIEC isolates. No DAEC strains were isolated from any of the samples examined. There was agreement between multiplex PCR and single PCRs assays for all strains. As demonstrated in Table 3, the multiplex PCR showed positive results for 68 diarrheagenic E.coli strains.The specificity of multiplex PCR was tested with the reference strains.Non-specific bands were not visualized(Figures 1, 2, 3).Two of theE.coli strains were positive for the ETEC lt marker plus additional virulence factors not typical of ETEC. One of the strains was positive for aggR, a known marker of EAEC. The second strain was positive for ipaH, which is EIEC marker.
The antibiotic susceptibility profile of the detected E.coli pathotypes is presented in Table 3. About 100% of the pathotypes were sensitive to Imipenem and Nitroforantoin, while susceptibility to other antibiotics was in the following order: Gentamycin 97.05%, Ciprofoxacin 88.23%, Nalidicsic acid 73.52%, Ceftazidime 66.66%, Chloramphenicol 64.70%. All of the pathotypes were 100% resistant to Ampicillin and Penicillin G, while 82.35%of the pathoypes were resistant to Trimethoprim-Sulphamethoxazol.
Disscussion
Phenotypic assays such as serogrouping with traditional antiserum are the routine methods widely used in clinical laboratories16, 17. Serotypic markers correlate, sometimes, extremely with specific categories of diarrheagenic E. coli;however, they are rarely enough in themselves to identify strains reliably as diarrheagenic.Thus, the finding of diarrheagenic E. coli has focused more and more on the identification of certain characteristicsbased on the presence of different chromosomal and/or plasmid encoded virulence genes that are not present in commensal E. coli9, 17.
Vaishnavi et al.,19reported that of 100 E. coli isolates only 25 were typeable and belonged to 14 different O-serogroups. They were 5 O153, 4 O102, 3 O25, 2 each of O130 and O169, and 1 each of O1, O8, O15, O37, O86, O101, O127, O143, and O160. In a study in Iran, the result of O-serogrouping by EPEC antisera showed that 17 (5.3%) isolates of the 321 E. coli isolates were typeable with the used antisera, and it was deduced that only 65% of the serogrouped EPEC strains carried virulence genes [10]. In a study in Kenya, it was shown that 76% of eaeA positive E. coli strains belonged to traditional EPEC serogroups19.
In the present study, the positive rates of pathogenic genes in the classical serogroup based E.coli strains were examined. According to our results and other studies, there was a relation between serogroup and pathogenic genes, but discovery of the virulence genes by PCR was more successful in detection of DEC. In this study,36.05% of the E.coli isolates were serogrouped by a latex agglutination test, while 46.25% of the isolates were detected to harbor virulence genes using PCR assays. It was also deduced that the association of stereotyping with PCR assay was statistically significant.
PCR analysis for identification of DEC groups has been designed to detect one or a few genes per reaction. However, multiplex PCRs are now being planned to detect multiple genes in the same reaction, thereby further reducing the cost and time required for the experimental procedure7, 20.In the present study, the direct challenge of standardizing three multiplex PCR assays is the probability for primer dimmers and nonspecific products. So, it is important to design primers with close annealing temperatures, and to use reference strains to find reaction specificity. Our results confirm that this technique can be administered for diagnosis of patients with the diarrhea, and the multiplex PCR assays showed high specificity for future epidemiological studies of DEC. This is an important observation from a public health point of view because clinical laboratories in Iran do not use molecular techniques for detection of DEC.
Table 1: Diagnostic polyvalent antisera in Sifin Kit.
| Serogroup | Antiserum |
| O26:K60; O44:K74; O114;K90; O125;K70; O142:K86; O158;K- | Anti-coli 1
|
| O55:K59; O86:K61; O91:K-; O111:K58; O119:K69; O126:K71; O127:K63; O128:K67
|
Anti-coli 2
|
| O25:K11; O78:K80; O103:K-; O118:K-; O124:K72; O145:K-; O157:K-; O164:K-
|
Anti-coli 3 |
Many PCR assays have been developed to identify the virulence genes of DEC. Tomaet al., [8]showed that it is possible to detect 11 of the major virulence genes of E. coli with four multiplex PCRs. For each of the target genes, various pairs of primers were selected from the studies.Also in a study by Hegde et al.,20two multiplex PCRs were standardized for identification of all DECs.They found that EAEC was the most common DEC usual in children with diarrheain India21. EAEC has also been reported as the leading DEC in children in studies carried out in Brazil22.EAEC has been reported to cause traveler’s diarrhea and persistent diarrhea in children in the developing countries with 19% reported in Goa and 33% in Guadalajara4, 15.These results are similar to our study, in which 13.7% of the E.coli isolates identified as EAEC. However, according to our study, with the exception of DAEC strain, which was not detected in 147 E.coli isolates, we were able to differentiate five categories of diarrheagenic E.coli, including the less common EIEC and variety of different serogroups of ETEC, EHEC and EPEC. The most frequent category of diarrheagenic E.coli detected was ETEC, which accounted for 17.8% of the isolates. This was followed by EAEC (13.7%); EHEC (7.5%), EPEC (6.2%) and EIEC (1.4%). These data corroborate the previously reported prevalence data, in which ETEC strains were amongst the commonest causes of intestinal disease in different parts of the world2, 23.Also there were two strains, which possessed lt plus ipaH and agg Rgenes, indicating that these two strains are a combination of ETEC-EAEC and ETEC-EIEC pathotypes. Some researchers believe that the virulence plasmid of EAEC and EIEC strains may have horizontally transferred to ETEC and EPEC recipient strains or vice versa3, 24.
Table 2: Primers used in the multiplex PCR for amplification of diarrhagenic E. coli genes
| Target organism | Target gene | Genom location | Primers(5′–3′) | Product size (bp) | |
| ETEC | st | Plasmid | ATT TTT ATT TCT GTA TTA TCT T C CAC CCG GTA CAT GCA GGA TT
|
190 | |
| lt | Plasmid | GGC GAC AGA TTA TAC CGT GC CGG TCTCTA TAT TCC CTG TT
|
450 | ||
| EPEC | eae
|
Chromosome | AGG CTT CGT CAC AGT TG CCA TCG TCA CCA GAG GA
|
570 | |
| bfp | Chromosome | AATGGTGCTTGCGCTTGCTGCGCCGCTTTATCCAACCTGGTA
|
326 | ||
| EHEC | stx1 | Phage | AGA GCG ATG TTA CGG TTT G AGA GCG ATG TTA CGG TTT G | 388
|
|
| stx2 | Phage | TGG GTT TTT CTT CGG TAT C GAC ATT CTG GTT GAC TCT CTT | 807 | ||
| EIEC | ipaH | Plasmid | GTT CCT TGA CCG CCT TTC CGA TAC CGT C 900 GCC GGT CAG CCA CCC TCT GAG AGT AC |
|
|
| EAEC | aggR | Plasmid | cta att gta caa tcg atg ta ga gtc cat ctc ttt gat aag
|
457 | |
| DAEC | daaE | Chromosome or plasmid | GAACGTTGGTTAATGTGGGGTAA TATTCACCGGTCGGTTATCAGT
|
542 | |
Tsukamoto and Kawai25 showed that the percentage of EPEC in the E. coli strains carrying eaeA was about 55% and according to Albert et al., 2657% of EAF-positive and/or eaeA positive EPEC strains belonged to a traditional serogroup. In the present study, the prevalence of tEPEC and aEPEC was 6.2%. The gene encoding eaeA was detected in 2.7% of the strains, and the bfp gene on the EAF plasmid was found only in 5 strains. According to our results, the rate of EPEC was similar to the study by Karam et al., in Iran,10but it was less than other reports (8.4%) from Iran27. Furthermore, only 5 of the EPEC strains were typeable with used antisera. One of the typical EPECs and 3 atypical EPEC were identified with antiserum 3, and only 1 atypical EPEC was positive with antiserum 2. In a study by Sunabe and Honma,28it was reported that there was no considerable difference between eae positive rates in the serogroup-based EPECs and EIECs. In the present study, the ipaH was detected in 2 of the E.coli isolates, and only one of the EIEC strains was serogrouped by aniserum 3. According to other studies,many of EHEC strains also have the eae gene; sopresence of stx genes can distinguish EHEC strains from EPEC strains19, 29.In our study, 11 EHEC strains were detected with the used primers. Seven of them had the stx2 gene, the frequent toxin phenotype pattern described in the Northern hemisphere. However, 4 strains harbored stx1 gene. It is to be mentioned that the prevalence and other geographical features of these pathogens as leading cause of diarrhea vary from country to country. The results of this study show that the validity of diagnosis with seroagglutination test needs to be addressed because O antigenic markers, sometimes, do not associate with pathogenicity of an isolate.
 |
Table 3: Antibiogram of E.colipathotypes isolated from the cases of diarrhea. |
Also, the E. coli found in this study expressed high levels of resistance to antimicrobials that are usually used in clinical medicine. This approves the fact that in most of the developing countries, diarrheal diseases are treated with insufficient regimen antibiotics and often without first identifying the pathogen4, 27. In Iran, the cost of materials needed to detect E. coli is about $5.00 per stool sample, and in this period of intensive care, if even one surgical procedure is obviated by the accurate diagnosis of E. coli infection or if even one severe secondary case of E. coli infection and colon cancer can be prevented, the cost of screening hundreds of stools for E. coli pathotypes can be justified 4.
 |
Table 4: Frequencies of serogroups associated with E.coli pathotypes.
|
Conclusion
In conclusion,differentiation between the diarrheagenic E. coli pathotypes is very important since they are involved in cancer or cellular senescence.So, this study helped to realize that DEC does contribute to the burden of diarrhea in people in the developing world [4, 15]. It was concluded that at attempts at economy are better conducted via finding appropriate criteria for requesting a stool culture rather than at limitation of the microbiological evaluation of specimens that are subjected.
Acknowledgment
The authors wish to thank Mrs. Firouzeh Safayeian for technical assistance and staff of the Microbiology Department of Tabriz University of Medical Sciences for providing facilities. We also thank the Drug Applied Research Centerof Tabriz University of Medical Sciences for the use of their equipments.
References
- Bonnet M, Buc E, Sauvanet P, Darcha C, Dubois D, Pereira B, Déchelotte P, Bonnet R, Pezet D, Darfeuille-Michaud A.Colonization of the human gut by E. coli and colorectal cancer risk. Clin Cancer Res, 2014; 15: 859-67.
- Vilchez S, Reyes D, Paniagua M, Bucardo F, Möllby R, Weintraub A. Prevalence of diarrhoeagenic Escherichia coli in children from Leon, Nicaragua. J Med Microbiol,2009;58: 630-637.
- Vidal M, Kruger E, Durán C, Lagos R, Levine M, Prado V, Toro C, Vidal R.. Single multiplex PCR assay to identify simultaneously the six categories of diarrheagenic Escherichia coli associated with enteric infections. J Clin Microbiol,2005;43: 5362-5365.
- Nataro JP. Enteroaggragative Escherichia coli pathogenesis. Curr Opin Gastroentrol, 2005; 21: 4-8.
- Guion CE, Ochoa TJ, Walker CM, Barletta F, Cleary TG. Detection of diarrheagenic Escherichia coli by use of melting-curve analysis and real-time multiplex PCR. J Clin Microbiol,2008;46: 1752-1757.
- March SB, Ratnam S. Latex agglutination test for detection of Escherichia coli serotype O157. JClin Microbiol,1989;27: 1675-1677.
- Aranda KR, Fabbricotti SH, Fagundes-Neto U, Scaletsky IC. Single multiplex assay to identify simultaneously enteropathogenic, enteroaggregative, enterotoxigenic, enteroinvasive and Shiga toxin-producingEscherichia coli strains in Brazilian children. FEMS Microbiol Lett,2007; 267: 145-150.
- Toma C, Lu Y, Higa N, Nakasone N, Chinen I, Baschkier A, Rivas M, Iwanaga M. Multiplex PCR assay for identification of human diarrheagenic Escherichia coli. J Clin Microbiol,2003;41: 2669-2671.
- Tamaki Y, Narimatsu H, Miyazato T, Nakasone N, Higa N, Toma C, Iwanaga M. The relationship between O-antigens and pathogenic genes of diarrhea-associated Escherichia coli. Jpn J Infect Dis, 2005; 8: 65-69.
- Karam MA, Bouzari S, Oloomi M, Aslani M, Jafari A. Phenotypic and genotypic characterization of Enteropathogenic Escherichia coli (EPEC) strains in Tehran, Iran. Iran J Microbiol,2010;2: 3-7.
- Nontongana N, Sibanda T, Ngwenya E, Okoh AI. Prevalence and antibiogram profiling of Escherichia coli pathotypes isolated from the Kat River and the Fort Beaufort abstraction water. Int J Environ Res Public Health,2014;11: 8213-8227.
- Peerayeh SN, Eslami M, Memariani M, Siadat SD. High Prevalence of blaCTX-M-1 Group Extended-Spectrum β-lactamase Genes in Escherichia coli Isolates From Tehran.Jundishapur J Microbiol2013; 7: 1-6.
- Oh KH, Park MS. Development of a One-Step PCR Assay with Nine Primer Pairs for the Detection of Five Diarrheagenic Escherichia coli Types. J Microbiol Biotechnol,2014; 24: 862-868.
- Chapman P. Evaluation of commercial latex slide test for identifying Escherichia coli O157. J Clin Pathol,1989; 42: 1109-1110.
- Samie A, Guerrant R, Barrett L, Bessong P, Igumbor E, Obi C. Prevalence of intestinal parasitic and bacterial pathogens in diarrhoeal and non-diarroeal human stools from Vhembe district, South Africa. J Health Popul Nutrition,2009; 27: 739-745.
- Moyo SJ, Maselle SY, Matee MI, Langeland N, Mylvaganam H. Identification of diarrheagenic Escherichia coli isolated from infants and children in Dar es Salaam, Tanzania. BMC Infect Dis,2007; 7: 92.
- Yang J-R, Wu F-T, Tsai J-L, Mu J-J, Lin L-F, Chen K-L, Kuo SH-S, Chiang C-S, Wu H-S. Comparison between O serotyping method and multiplex real-time PCR to identify diarrheagenic Escherichia coli in Taiwan. J Clin Microbiol,2007; 45: 3620-3625.
- Vaishnavi C, Kaur S, Beutin L, Krueger U. Phenotypic and molecular characterization of clinically isolated Escherichia coli. Indian J Pathol Microbiol,2010; 53: 503-508.
- Nagayama K, Yamada K, Yamamoto K, Honda T. Detection of bundle-forming pili (BFP) gene and attaching and effacing (eae) gene in E. coli isolated from patients in Kenya. Kansenshogaku Zasshi,1996;70: 142-146.
- Pass M, Odedra R, and Batt R. Multiplex PCRs for Identification of Escherichia coli Virulence Genes.J Clin Microbiol,2000; 38: 2001-2004.
- Hegde A, BallalM, Shenoy S. Detection of diarrheagenic Escherichia coli by multiplex PCR.Indian J Med Microbiol,2012;30: 279-284.
- Regua-Mangia A, Gomes T, Vieira M, Andrade J, Irino K, Teixeira L. Frequency and characteristics of diarrhoeagenic Escherichia coli strains isolated from children with and without diarrhoea in Rio de Janeiro, Brazil. J Infect,2004; 48: 161-167.
- Gomez-Duarte OG, Romero-Herazo YC, Paez-Canro CZ, Eslava-Schmalbach JH, Arzuza O. Enterotoxigenic Escherichia coli associated with childhood diarrhoea in Colombia, South America. J Infect Dev Ctries,2013;7: 372-381.
- Taniuchi M, Walters CC, Gratz J, Maro A, Kumburu H, Serichantalergs O, Sethabutr O, Bodhidatta L, Kibiki G, Toney DM. Development of a multiplex polymerase chain reaction assay for diarrheagenic Escherichia coli and Shigella spp. and its evaluation on colonies, culture broths, and stool. Diagn Microbiol Infect Dis,2012;73: 121-128.
- Tsukamoto T, Kawai T. The eae gene, adherence to HeLa cells and serotypes of Escherichia coli isolated from diarrhea.Kansenshogaku Zasshi,1995; 69: 85-90.
- Albert MJ, Faruque SM, Faruque ASG, Neogi PKB, AnsaruzzamanM, Bhuiyan NA, Alam K, Akbar MS. Controlled study of Escherichia coli diarrheal infections in Bangladeshi children. J Clin Microbiol,1995; 33: 973-977.
- Jafari F, Shokrzadeh L, Hamidian M, Salmanzadeh-Ahrabi S, Zali MR. Acute diarrhea due to enteropathogenic bacteria in patients at hospitals in Tehran. Jpn J Infect Dis,2008; 61: 269-273.
- Sunabe T, Honma Y. Relationship between O-Serogroup and Presence of Pathogenic Factor Genes in Escherichia coli.Microbiol Immunol,1998; 42:845-349.
- Blanco M, Blanco JE, Dahbi G, Alonso MP, Gutierrez AM, Coira MA, Madrid C, Juárez A, Bernárdez MI, González EA. Identification of two new intimin types in atypical enteropathogenic” Escherichia coli“. Int microbial,2006;9: 103-110.

This work is licensed under a Creative Commons Attribution 4.0 International License.





