Manuscript accepted on : 25 March 2012
Published online on: --
Cloning and Sequence Analysis of Lipase Gene from DMS3 Isolate
M. P. Widhiastuty1, Setyanto Tri Wahyudi1,2, Maelita Ramdani Moeis3, Fida Madayanti1 and Akhmaloka1*
1Biochemistry Research Group, Faculty of Mathematics and Natural Sciences, Institut Teknologi Bandung, Jln Ganesha 10, Bandung, Indonesia. 2Biophysics Division, Department of Physics, Bogor Agricultural University, Bogor, Indonesia. 3Microbiology, Genetics and Molecular Biology Research Group, School of Life Sciences and Technology, Institut Teknologi Bandung, Jln Ganesha 10, Bandung, Indonesia.
ABSTRACT: Genes encoding a lipase from DMS3, isolated from Kawah Domas hotspring, Indonesia was cloned and characterised. The lipase gene was cloned based on PCR amplification from genomic DNA. Two set of primers was designed to amplify the lipase gene. An open reading frame of 1248 bps encoding polypeptide of 416 amino acid residues, has been amplified and sequenced. The sequence of amplicon showed high homology (99%) with lipase from Geobacillus thermoleovorans. Detail comparison among three highest homology of lipase showed that there are some variation of amino acid residues. However the subtitution of amino acid residue in lipase ITB3.1, especially at amino acid residue 194 from Asp to Asn is predicted to showh no significant different for the characteristic of lipase ITB3.1 compared to that the other lipase from family 1.5.
KEYWORDS: Lipase; Kawah Domas Hot Spring; PCR Cloning; Geobacillus thermoleovoran
Download this article as:| Copy the following to cite this article: Widhiastuty M. P, Wahyudi S. T, Moeis M. R, Madayanti F, Akhmaloka. Cloning and Sequence Analysis of Lipase Gene from DMS3 Isolate. Biosci Biotech Res Asia 2012;9(1) |
| Copy the following to cite this URL: Widhiastuty M. P, Wahyudi S. T, Moeis M. R, Madayanti F, Akhmaloka. Cloning and Sequence Analysis of Lipase Gene from DMS3 Isolate. Biosci Biotech Res Asia 2012;9(1). Available from: https://www.biotech-asia.org/?p=9532 |
Introduction
Lipases (triacyglycerol acylhydrolases, EC 3.1.1.3) catalyze the hydrolysis of the ester bonds of long-chain triglycerides into fatty acids and glycerol. These enzymes are particular importance in biotechnology because of diverse applications such as on detergents, food production, pharmaceuticals and synthesis of fine chemical industries1,2,3,4.
Lipase from thermophiles have became objects of interest for understanding of basic properties and industrial applications since they exhibit higher thermodynamic stability at elevated temperatures and in organic solvents5. Enzyme from an organism usually has temperatur or pH optimum at or close to its growth condition as consequence of adaptation of the corresponding microorganism to its enviroment5,6. Some thermostable lipase have been isolated from Bacillus and Geobacillus. Most of them showed optimum activity at temperature above 55oC and stable in various organic solvent7,8,9. The immobilized lipase from Bacillus stearothermophilus MC7 even exhibits good operational thermostability with half-life of 50 days at 60oC in a solvent-free system10.
Previously, we reported several thermophilic microorganisms from various sources in Indonesia, including some hot springs and thermogenic compost11,12,13,14. Some of the microorganisms showed lipolytic activity, and most of them were identified as member of genera Bacillus and Geobacillus based on its 16S rRNA gene sequences15. Among them, DMS3 isolate has the highest lipolytic activity. In this report we describe cloning and characterisation of lipase gene from DMS3 isolate. The cloning was carried out based on PCR methods and the characterisation of the gene was based on its nucleotide sequences.
Materials and Methods
Materials
Peptone, yeast extract, NaCl, Tris base, EDTA and potassium acetate were supplied from Biobasic. Lysozyme and proteinase were purchased from Fermentas. HCl, acetic acid glacial, chloroform, isoamyl alcohol and ethanol were purchased from Sigma. All other reagents were analytical grade unless otherwise stated. Primer Flip1, Flip2, Rlip1 and Rlip2 were used to amplify the gene (Table 1). DMS3 isolate was taken from microbe collections in our laboratory. DMS3 originally was isolated from Kawah Domas Hot Springs, West Java, Indonesia.
Cultivation of Microbes
The bacterial cultures were cultivated in media ½ T (pepton 0.4%, yeast extract 0.2%, NaCl 0.1%) and incubated at 70oC for 18 hours. The pellet cell was collected by centrifugation at 6000 g for 10 minutes.
Isolation of Chromosomal DNA
The pellet cells were suspended in 10 mM Tris HCl buffer (pH 8.0) containing 8 mg/ml of lysozyme and incubated at 37 °C for 1 h. The cells were lysed by adding lysis buffer containing 2% SDS, 0.8 mg/ml proteinase K and 200 mM EDTA pH 8.0. The lysis process was carried out by incubation at 50 oC for 30 min. Ice cold potassium acetate and acetic acid glacial mixed solution were added. The denatured proteins were precipitated by centrifugation at 8000 g for 15 min. Supernatants were mixed with an equal volume of chloroform isoamylalcohol (24:1, v/v). The aqueous phase was recovered by centrifugation and precipitated with 0.6 volume of isopropanol at room temperature for 1 h. The pellet of crude nucleic acids were obtained by centrifugation at 16.000 g for 20 min at room temperature, washed twice with cold 70% ethanol, and re-suspended in sterile deionized water.
Table 1: Primers used for amplification of lipase gene
| Primer | Sequences |
| Flip1 | 5’- CACCCATCGTGCTTCTCCAT -3’ |
| Flip2 | 5’- ATGTGAGGGGAGGAGAAGG-3’ |
| Rlip1 | 5’- CCCTTGGCTGTGGGCGA -3’ |
| Rlip2 | 5’- GAGCCATCCGATCGAGATG -3’ |
Table 2: Homology analysis of lipase ITB3.1 with lipase from various microorganism.
| No | Accession No. | Description | %
homology |
Score |
| 1 | AF134840_1 | Geobacillus thermoleovorans | 99 | 858 |
| 2 | AAX11388.1 | Geobacillus stearothermophilus | 99 | 855 |
| 3 | ADU94368.1 | Geobacillus sp. Y412MC61 | 99 | 855 |
| 4 | ADI26534.1 | Geobacillus sp. C56-T3 | 98 | 822 |
| 5 | AAO92067.2 | Geobacillus zalihae | 98 | 821 |
| 6 | JC8061 | Geobacillus sp. T1 | 97 | 820 |
| 7 | AAW47928.1 | Bacillus sp. L2 | 98 | 820 |
| 8 | AAV35102.1 | Bacillus sp. 42 | 98 | 819 |
| 9 | AAY82869.1 | Geobacillus sp. SF1 | 96 | 810 |
| 10 | AAM21775.1 | Bacillus sp. Tosh | 96 | 809 |
| 11 | ABC48693.1 | Geobacillus thermoleovorans | 95 | 807 |
| 12 | AAF40217.1 | Geobacillus stearothermophilus | 96 | 805 |
| 13 | CAL36912.1 | Geobacillus thermoleovorans | 98 | 804 |
| 14 | AAC12257.1 | Geobacillus stearothermophilus | 92 | 801 |
| 15 | AAM21774.1 | Geobacillus thermoleovorans | 95 | 800 |
| 16 | ACJ07039.1 | uncultured bacterium | 92 | 798 |
| 17 | BAH28804.1 | Geobacillus sp. SBS-4S | 99 | 796 |
| 18 | BAD76271.1 | Geobacillus kaustophilus HTA426 | 95 | 796 |
| 19 | AEB71527.1 | Geobacillus sp. MNK | 93 | 782 |
| 20 | CAA64621.1 | Geobacillus thermocatenulatus | 95 | 781 |
| 21 | ACN79581.1 | Geobacillus sp. NTU 03 | 92 | 777 |
| 22 | AF429311_1 | Geobacillus stearothermophilus | 95 | 776 |
| 23 | ABK34427.1 | Geobacillus stearothermophilus | 95 | 773 |
| 24 | AF141874_1 | Bacillus sp. TP10A.1 | 93 | 759 |
| 25 | ACS93141.1 | Geobacillus sp. RD-2 | 95 | 748 |
Table 3: Comparation of amino acid residues between Lipase ITB3.1 and G. thermoleovorans, G. stearothermophilus and Geobacillus sp lipases with accession number AF134840.1, AAX11388.1, ADU94368.1 respectively.
| No residues | DMS3 | G. thermoleovorans | G. stearothermophilus | Geobacillus sp. |
| 3 | Cys | Cys | Gly | cys |
| 12 | Gly | Gly | Gly | Gln |
| 23 | Pro | Ser | Ser | Pro |
| 31 | Val | Ala | Thr | Ala |
| 194 | Asn | Asp | Asp | Asp |
| 225 | val | val | Val | Ala |
| 370 | Ala | Ala | Ala | Val |
| 416 | Arg | Arg | Gln | Gln |
Amplification of lipase gene
Amplification of lipase gene was carried out by PCR method. Flip1 and Rlip1 were used as internal primers, while Flip2 and Rlip2 were used as external primers. A total of 50 µL reaction mixture consists of 5 ng DNA template, 10 pmol of primers, 200 µM dNTP mixture, 5 µL 10x PCR buffer, 1,25 U Taq Polymerase, was used for PCR reaction. The PCR were carried out using i-Cycler (Bio-Rad). The reaction conditions used were: 1 cycle (95oC for 5 min), 30 cycles (95oC for 1 min, 50oC 1 min, and 72oC for 2 min), and final cycle of 72oC 10 min.
Homological analysis of Lipase Gen Sequence
Sequences of the full length PCR amplicon were obtained by primer walking strategy and assembling of the partial sequences using the tool SeqManTM of the software packet DNA-Star. Sequence homology analysis of the lipase was carried out by comparing the nucleotide sequence of local lipase genes with nucleotide sequence from the Gene Bank database at NCBI (National Centre of Biotechnological Information) through web site http://www.ncbi.nlm.nih.gov using BLAST program for screening of sequence similarity. Sequences alignments were performed by ClustalX program16 and visualized using GenDoc program.
Structure modelling
The initial coordinate of Lipase ITB1.2 was obtained from homology structure modelling using Swiss-Model17. Three dimensional structure visualization of Lipase ITB3.1 was generated using Visual Molecular Dynamics (VMD) software version 1.918.
Nucleotide sequence accession numbers
Nucleotida sequence of lipase gene for DMS3 isolate has been deposited in the GenBank database under accession number HQ398859.
Result
Cloning of Lipase ITB3.1
The culture of DMS3 showed high lipase activity15. The lipase gene from DMS3 isolate was cloned based on PCR method directly from genomic DNA. The critical step in the amplification of gene from the genomic DNA is on designing and choosing the primers, especially if the genes have only few conserved of amino acids in its sequences19. Two conserved region in lipase gene are often used in designing primers19, one region was at around one of three lipase’s catalytic residues, the amino acid serin. This region is known as conserved pentapeptide which always in form of Gly-X-Ser-X-Gly20. Another region is the oxyanion hole region which located as 60-108 aa upstream of the conserved pentapeptide19. The region is recognized by the presence of short hydrophobic region (6 aa) upstream of moderately conserved His-Gly (HG) dipeptide. In this study, a set of internal primers were designed based on sequence in these two conserved regions. The primers were used to amplify fragment of lipase gene from DMS3 isolate with length of approximately 300 bp (Fig. 1A). The nucleotide sequence of thisfragment, aligned with sequence data in GenBank, showed high homology with all sequence of lipase gene from Geobacillus, indicating that DMS3 isolate was belong to genus Geobacillus. The homology result was in agreement with identification of the DMS3 isolate based on its 16S rRNA gene sequence15. In order to amplify the whole gene of lipase, another pair of primer, external primers, were further designed. External primers were designed based on conserved region upstream and downstream of coding region. 6 lipase genes which showed closest homology with the DMS3 lipase were used for designing external primer. The external primer amplify polinucleotide with length of approximetly 1300 bp (Fig. 1B). The gene was further named as lipase ITB3.1.
 |
Figure 1: Agarose gel electrophoregram of PCR (A) Fragment of lipase gene as result of amplification using internal primer (lane 1). (B) Lipase gene of DMS3 isolate (lane 4). Lane 2 & 3 Marker PUC19/Hinf I.
|
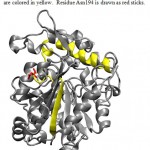 |
Figure 2: 3D Structure of Lipase ITB3.1. b-6 and a-6 are colored in yellow. Residue Asn194 is drawn as red sticks.
|
Homological analysis of Lipase ITB3.1 sequences
The complete sequence of lipase ITB3.1 consists of 1248 nucleotide sequences (gene bank accession number HQ398859). Homological analisis of 416 deduced amino acid sequences using blastN program through website http://ncbi.nlm.nih.gov showed that lipase ITB3.1 has high homology with lipase from Geobacillus. The result suggested that lipase ITB3.1 is one of the member of bacterial lipase group family 1.5. Table 2 shows 50 lipases which were close to lipase ITB3.1. The highest homology of lipase ITB3.1 was lipase from G. thermoleovorans (accession number AF134840) with 3 aa differences. Detail comparison of lipase ITB3.1 to the three best homologs (G. thermoleovorans, G. stearothermophilus and Geobacillus sp. lipases with accession number AF134840.1, AAX11388.1, ADU94368.1 respectively) showed that there are few variation of amino acid residues among them (Table 3).
The three dimensional structure of Lipase ITB1.2 was constructed by homology modelling with Swiss-Model and the result showed highest homology (98,7%) with Lipase T1 (2DSN.PDB) from G. zalihae.
Discussion
Lipase ITB3.1 showed highest homology to lipase from G. thermoleovorans (accession number AF134840.1). Based on 16S rRNA gene sequence previously reported, DMS3 isolate was closely related to G. kaustophillus (accession number EU652092) and G. thermoleovorans (acession number AY074879), both of them located in the same branch. However, homological analysis of lipase ITB3.1 with lipase from G. kaustophillus HTA426 was only 95% compared to that homology of the gene to lipase from G. thermoleovorans which was 99%. From the homology analysis of lipase ITB3.1 and 16S rRNA gene sequences of DMS3 isolate, suggested that DMS3 isolate show closer to G. thermoleovorans compared to that the G. kaustophillus.
Lipase ITB3.1 shows 99% homology with three published lipases and at least 92% homology with other lipases from sub-family 1.5 (Table 2). The lipase has same catalytic residues in the same position with other lipases from sub-family 1.5. There are no differences in amino acid residues observed in oxyanion hole as well as the lid. Lipase ITB3.1 has 8 amino acid differences compared to three closest related lipase from G. thermoleovorans, G. stearothermophilus and Geobacillus sp.with accession number AF134840.1, AAX11388.1, ADU94368.1 respectively. Among them, one residue (Asn194) which is conserved among three other lipases but substituted on lipase ITB3.1 (Table 3). The residue of Asn194 was replacing residue Asp194 on other lipases. Alignment analysis of amino acid sequences of lipase ITB3.1 and other lipases showed residue Asn194 probably located in a turn between b-6 and a-621. The a-6 and a-7 were identified as lid in lipase sub-family 1.522. Asn194 residue probably located in the surface of the enzyme, which has contact with solvent, as shown in Fig 2. Therefore the substitution of Asp to Asn which is a negatively to neutrally charged amino acid may not exhibit significant effect to the characteristic of lipase ITB3.1.
Refrences
- Krishna S.H, dan Karanth N.G. Lipases and Lipase-Catalyzed Esterification Reactions in Nonaqueous Media. Catalysis Reviews, 2002; 44 (4): 499 – 591.
- Sharma, R., Chisti, Y., dan Banerjee, U. C. Production, Purification, Characterization, and Application of Lipase, Biotechnol Adv., 2001; 19: 627 – 662.
- Jaeger, K. E., dan Eggert, T. Lipases for Biotechnology, Current Opinion in Biotechnology, 2002; 13: 390-397
- Jegannathan, K. R. dan Abang, S. Production of Biodiesel Using Immobilized Lipase-A Critical Review, Critical Reviews in Biotechnology, 2008; 28: 253-264.
- Sinchaikul, S., Sookkheo, B., Phutrakul, S., Pan, F. P., dan Chen, S. T. Optimization of a Thermostable Lipase from Bacillus stearothermophilus P1: Over-expression, Purification, and Characterization, Protein Expression and Purification, 2001; 22: 388–398.
- Yano, J.K., dan Poulos, T. L. New Understandings of Thermostable and Peizostable Enzymes, Current Opinion in Biotechnology, 2003; 14: 360–365
- Abdel-Fattah, Y. R., dan Gaballa, A. A. Identification and Over-Expression of A Thermostable Lipase from Geobacillus thermoleovorans Toshki in Escherichia coli, Microbiological Research, 2008; 163: 13-20.
- Leow, T. C., Zaliha, R.N., Rahman, R.A., Basri, M., dan Salleh, A.B. A Thermoalkaliphilic Lipase of Geobacillus sp. T1, Extremophiles, 2007; 11: 527–535.
- Kumar, S., Kikon, K., Upadhyay, A., Kanwar, S.S., dan Gupta, R. Production, Purification, and Characterization of Lipase from Thermophilic and Alkaliphilic Bacillus coagulans BTS-3., Protein Expression and Purification, 2005; 41: 38-44.
- Guncheva, M. D., Zhiryakova, D. dan Radchenkova, N. Acidolysis of Tripalmitin with Oleic Acid Catalyzed by a Newly Isolated Thermostable Lipase, Am. Oil Chem. Soc., 2008; 85: 129 – 132.
- Yohandini H., F. Madayanti, P. Aditiawati, dan Akhmaloka. Diversity of Microbiol Thermophiles in a Neutral Hot Spring (Kawah Hujan A) of Kamojang Geothermal Field, Indonesia, Journal of Pure and Applied Microbiology, 2008; 2:
- Aminin, A. L. N., M. Asy’ari, N. S. Mulyani, F. Madayanti, P. Aditiawati, dan Akhmaloka. 16S Ribosomal RNA-Based Analysis of Thermophilic Bacteria in Gedongsongo Hot Spring. Indonesian Journal for Microbiology, 2006; 1 (1): 37-42.
- Akhmaloka, Pramono H, Ambarsari, L., Susanti, E., Nurbaiti, S., dan Madayanti, F. Cloning, Homological Aanalysis, and Expression of DNA Pol I from Geobacillus thermoleovorans, International Journal of Integrative Biology, 2007; 1 (3): 206-215.
- Madayanti F. W., Viera E., Widhiastuty, M. P., dan Akhmaloka. Characterization and Identification of Tthermophilic Lipase Producing Bacteria from Thermogenic Compost, Journal of Pure & Applied Microbiology, 2008; 2 (2): 325-332.
- Widhiastuty, M.P., Madayanti, F., Moeis, M.R., dan Akhmaloka. Characterization and Identification of Thermostable Alkaline Lipase Producing Bacteria from Hot Spring around West Java, Journal of Pure and Applied Microbiology, 2009; 3 (1): p 27-40
- Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, dan D. J. Lipman. Gapped BLAST and PSI-BLAST: a New Generation of Protein Database Search Programs, Nucleic Acids Res., 1997; 25 (17): 3389-3402
- Arnold K., Bordoli L., Kopp J., and Schwede T. The SWISS-MODEL Workspace: A web-based environment for protein structure homology modelling. Bioinformatics, 2006; 22: 195-201.
- Humphrey, W., A. Dalke, & K. Schulten. VMD – Visual molecular dynamics. Molec. Graphics. 1996; 14 (1): 33-38.
- Bell, P. J. L., Sunna, H., Gibbs, M. D., Curach, N. C., Nevalainen, H., dan Bergquist, P. L. Prospecting for novel lipase genes using PCR, Microbiology, 2002; 148: 2283–2291.
- Jaeger, K. E., Dijkstra, B. W., Reetz, M. T. Bacterial Biocatalysts: Molecular Biology, Three-Dimensional Structures, and Biotechnology Applications of Lipases, Rev. Microbiol., 1999; 53: 315-351
- Jeong, S. T, Kim, H., Kim, S., Chi, S., Pan, J., Oh, T., dan Ryu, S. Novel Zinc-Binding Center and a Temperature Switch in The Bacillus stearothermophilus L1 Lipase, J Biol Chem., 2002; 277: 17041-17047.
- Carrasco-Lopez, C., Godoy C, Rivas, B., Fernandez-Lorente, G., Palomo, J.M., Guisan, J.M., Fernandez-Lafuente, R., Martinez-Ripoll, M., dan Hermoso, J.A. Activation of Bacterial Thermophilic Lipase is Spurred by Dramatic Structural Rearrangements, Journal of Biological Chemistry, 2009; 284 (7): 4365-4372.

This work is licensed under a Creative Commons Attribution 4.0 International License.





