Manuscript accepted on : May 05, 2010
Published online on: 28-12-2010
Isolation And Identification Of Genus Lactobacillus From Different Curd Samples
Renuka Goyal and Harish Dhingra
Department of Science, FASC, Mody Institute of Technology and Science (Deemed University), Lakshmangarh - 332 311 India.
Corresponding Author E-mail:renukagoyal@ymail.com,
ABSTRACT: Lactobacillus is a genus of lactic acid bacteria. Lactic acid bacteria mainly found in fermented dairy products. This study was carried out for the identification and characterization of Lactobacillus isolated from curd. A total of 14 curd samples were collected from different places of Gurgaon (Haryana) and Lakshmangarh (Rajasthan). Their identification was carried out based on their morphological and biochemical characteristics. A total of 28 isolates were screened on the basis of their phenotypic characteristics. Lactobacillus showed cream-white colonies, rod-shaped, gram-positive, non-spore forming, non-motile, and catalase negative bacteria. Some of the Lactobacillus species are also showing the characteristics of homofermentative or heterofermentative categories.
KEYWORDS: F-Fermentation: H-Homofermentation (production of lactic acid but not gas)
Download this article as:| Copy the following to cite this article: Goyal R, Dhingra H. Isolation And Identification Of Genus Lactobacillus From Different Curd Samples. Biosci Biotech Res Asia 2010;7(2) |
| Copy the following to cite this URL: Goyal R, Dhingra H. Isolation And Identification Of Genus Lactobacillus From Different Curd Samples. Biosci Biotech Res Asia 2010;7(2). Available from:https://www.biotech-asia.org/? p=9450 |
Introduction
Lactobacillus is highly used in controlled fermentation. Lactic acid bacteria widely used in traditional fermented milk, in industrial, fermentation process and starter culture in dairy industry. Lactic acid bacterial starter cultures are used to produce lactic acid at a suitable rate for the fermentation process. The primary function of bacterial culture is to convert the lactose into glucose or galactose, and then these sugars are converted to the end product of lactic acid. Now a day’s lactic acid is being used by the food industry as acidulent and preservative for the production of cheese and yoghurt.
Genus Lactobacillus isolated from curd shows absence of catalase activity. The genus produces lactic acid during fermentation of glucose which can be detected by Hugh and Leifson’s test.
The objective of the present study was aimed to identify and characterised the strains of lactic acid bacteria isolated from the curd samples on the basis of their phenotypic characterization.
Materials and Methods
Sample collection
A total of 14 samples of curd were collected from different places of Gurgaon (Haryana) and Lakshmangarh (Rajasthan) and stored at 4ºC. Then all the samples were taken to the Biotechnology and Microbiology Laboratory of MITS (Deemed University), Lakshmangarh for further microbiological analysis.
Isolation of the Bacteria
Selective media for lactic acid bacteria is MRS (de Man, Rogosa and Sharpe) agar media. A loopful of the samples was streaked on the sterile MRS agar petri plate by quadrant streaking method, under aseptic conditions. After streaking all the petri plates were incubated at 37ºC for 24-48 hours. Then from these plates isolated colonies were restreaked on the MRS agar slants and then stored at 4ºC.
Phenotypic Characterization
Identification of the isolates was performed on the basis of their morphological and biochemical characteristics as followed:
Gram Staining- Initially identification of the isolates was done by gram staining method (Hans Christian Gram, 1884).
Microscopic Morphology
Gram staining slides were observed under microscope oil immersion.
Endospore Test
Bacterial smear was made on microscopic slide under aseptic conditions and heat fixed. Then slide was placed over the steaming water bath and malachite green (primary stain) was applied for 5 minutes. Slide was removed from the water bath and rinsed with water until water run clear. Then the slide was flooded with the counter stain safranin for 20 seconds and rinsed with water. Then slides were blot dried and observed under the light microscope.
Hugh and Leifson’s test
The purpose of this test was to determine whether an organism is an oxidizer or a fermenter on the basis of production of acid in aerobic and anaerobic conditions. Hugh and Leifson’s medium was prepared into culture tubes. All these test tubes were autoclaved at 121ºC for 15 minutes. A syringe filter sterilized solution of 10% carbohydrate (glucose) was aseptically added to the medium to a final concentration of 1%. Medium was cooled and inoculated by stabbing with the test organism. After stabbing, all the culture tubes were kept in incubator under aerobic and anaerobic conditions at 37ºC for 24-48 hours. After incubation, all the test tubes were observed for fermentation.
Motility test
Hugh and Leifson’s medium was also used for the testing that the bacteria were motile or non-motile through stab inoculation.
Catalase test
This test was used to check the production of enzyme catalase. For this test a clean microscopic slide was taken. A drop of 3% H2O2 was taken on the slide aseptically. A loopful of bacterial culture was taken and mixed with 3% H2O2 solution on the slide and the presence of the bubble production was observed.
Sugar Fermentation test
Approximate 100 ml of the nutrient broth solution was prepared in conical flask and 1 ml phenol red was added to it. This medium was autoclaved at 121ºC for 15 minutes and cooled at room temperature. A syringe filter sterilized solution of 1% glucose was prepared under aseptic conditions.
In all sterilized test tube 5ml of the broth and 100µl of the glucose solution was taken and labelled. All the test tubes were kept at room temperature for 24 hours to check the contamination. After 24 hours, all these test tubes were inoculated with freshly grown bacterial culture and incubated at 37ºC for 48 hrs.
In case of homofermentation colour changes from red to yellow. And in heterofermentation there is gas production in Durham tube along with the change in the colour.
Table 1: Identification of bacteria.
| S.No. | Isolates | Types of colonies | Gram staining | Microscopic morphology | Endospore test | Hugh and leifson’s test | Motility test | Catalase test | Sugar fermentation test |
| 1
2
3
4
5
6
7
8
9
10
11
12
13
14 |
S1
L1 S2 L2 S3 L3 S4 L4 S5 L5 S6 L6 S7 L7 S8 L8 S9 L9 S10 L10 S11 L11 S12 L12 S13 L13 S14 L14 |
Small
Large Small Large Small Large Small Large Small Large Small Large Small Large Small Large Small Large Small Large Small Large Small Large Small Large Small Large |
+
+ + + + + + + + + + + + + + + + + + + + + + + + + + + |
Rod shaped
Rod shaped Rod shaped Rod shaped Rod shaped Rod shaped Rod shaped Rod shaped Rod shaped Rod shaped Rod shaped Cocci Cocci Cocci Rod shaped Rod shaped Rod shaped Rod shaped Cocci Rod shaped Rod shaped Rod shaped Rod shaped Rod shaped Rod shaped Rod shaped Cocci Rod shaped |
–
– – – – – – – – – – – – – – – – – – – – – – – – – – – |
F
F F F F F F F F F F F F F F F F F F F F F F F F F F F |
–
– – – – – – – – – – – – – – – – – – – – – – – – – – – |
+
+ + + + + + – – + + – + + + + + – + + + + – + – – + + |
H
H H H H H H H H H H H H H H H H H H H H H H H H H H H |
Results
A total of 28 different isolates were isolated from 14 different curd samples after streaking a loopful of curd sample on the MRS agar plate by quadrant streaking method. Two different types of small and large creamy to white coloured colonies were obtained.
Gram staining of the bacterial culture showed they were gram positive and their cell morphology was rod shaped and some of them were coccoid shaped.
Endospore test showed that the bacteria were non-spore forming, showing negative result (red colour) instead of forming positive result (green colour).
Hugh and Leifson’s test showed that all the bacterial cultures were capable of producing fermentation. Fermentative organisms developed acidic conditions in anaerobic conditions, while oxidative organisms produced acid in aerobic conditions (fig: 1).
 |
Figure 1: Hugh and Leifson’s test.
|
And when this medium culture tubes were used to determine the motility of bacteria, then it was found that the bacteria were non-motile, growing in a confined stab line (fig: 3) instead of making the whole medium turbid.
Catalase test showed that the only 7 strains out of 28 strains did not produce bubbling when mixed with3% H2O2 this showed that there was absence of catalase enzyme. The absence of catalyse enzyme showed that identified bacteria was from Lactobacillus species.
Further, if these cultures were used for sugar fermentation showed that the bacteria was of homofermentative nature by changing colour of the medium (fig: 2) from red to yellow because of acid production.
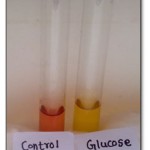 |
Figure 2: Sugar Fermentation test.
|
Discussion
Mehmood et al (2009) isolated Lactobacillus from dairy samples. All the isolates were gram positive, rod shaped, non- spore forming and showing fermentation and the absence of catalase enzyme. In our present investigation a total of 28 isolates from curd were screened for various characteristics and it was found that the all isolates were gram positive , rod shaped, non-spore forming, and a few of them were catalase negative.
Catalase test was used for the detection of the enzyme catalase which has been involved in conversion of H2O2 into H2O and O2. A positive result was screened by the presence of the bubble (O2), indicated that bacteria survive under aerobic conditions. During aerobic respiration bacteria produce hydrogen peroxides, a natural by product of metabolism. In our present investigation few of strains were showing the absence of catalase enzyme.
Forouhandeh et al (2010) and Mehmood et al (2009) determined that during sugar fermentation test, Lactobacillus produce acid from glucose while gas was not produced. In our investigation acid production was evidence by change in colour of sugar from red to yellow. However there was no gas production in Durham tube.
Ahmed and Kanwal (2004) isolated different strains of Lactobacillus from camel milk and reported that all the strains were non-motile. In our present study it was found that the strains which we isolated from curd samples were also showing the characteristics of non-motile nature.
Coeuret et al (2003) isolated, characterised, and identified the Lactobacilli mainly from cheese and other dairy products and reported that Lactobacilli species were non-spore forming. In our results all the strains were non-spore forming.
Nair and Surendran (2005) isolated lactic acid bacteria from various samples of fresh and frozen fish and prawn. All the cultures were identified on the basis of their morphological, cultural, physiological and biochemical characteristics. For Hugh and Leifson’s test oxidation-fermentation medium used for evaluating the production of acid and gas from 1% glucose. And reported that some of the strains were showing positive result and some were negative result. In our isolates all of the strains were showing the positive result with the production of acid.
 |
Figure 3: Motility test.
|
Conclusion
A total of 28 different strains were isolated from 14 different curd samples were performed. All the strains were identified on the basis of their colonies morphology and other biochemical characteristics. And it was found that all the strains were non-motile, non-spore forming, Gram-positive, rod shaped bacteria that produce lactic acid homofermentatively from glucose. Few strains were showed the absence of catalase enzyme because bacteria use peroxidase to detoxify H2O2, an enzyme that does not evolve O2.
It was concluded that we successfully isolated Lactobacillus from curd samples by using a range of Lactobacillus- specific morphological and biochemical test to determine the presence of Lactobacillus in the isolates. In our investigation we could not isolates Lactobacillus from all the curd samples. Only out of 28 isolates, seven isolates will show positive test for Lactobacillus.
Acknowledgement
We thank Dr. R.K.Gaur, Head of the Department of Biotechnology and Microbiology and Dean Dr. Shakti Baijal, MITS Lakshmangarh (Deemed University) for their support in completing the work.
References
- Abid R., Zeinoddin S.M., and Zad S.S., Identification of lactic acid bacteria isolated from traditional Iranian Lighvan cheese. Pakistan Journal of Biological Sciences 9(1):99-103 (2006).
- Ahmed T. and Kanwal R., Biochemical characteristics of lactic acid producing bacteria and preparation of camel milk cheese by using starter culture. Pakistan Vet. J., 24(2) (2004).
- Amanullah S.M., Alam M.S., Subarna R.N., Bateen R., Huque S.K. and Sultana A., Feeding Lactobacilli as probiotic and proportion of Escherichia coli in the intestine of calves. The Bangladesh Veterinarian 26 (1):17-22 (2009).
- Coeuret V., Dubernet S., Bernardeau M., Gueguen M., and Vernoux J.P., Isolation, characterization and identification of Lactobacilli focusing mainly on cheese and other dairy products. Lait 83, 269-306 (2003).
- Amin M., Jorfi M., Khosravi A.D., Samarbafzadeh A.R., Sheikh F.A., Isolation and identification of Lactobacillus casei and Lactobacillus plantarum from plant by PCR and detection of their antibacterial activity. Journal of Biological Science 9(8):810-814 (2009).
- Zahoor T., Rahman S.U., and Farooq U., Viability of Lactobacillus bulgaricus as yoghurt culture under different preservations methods. International Journal of Agriculture & Biology 05:46-48 (2003).
- Erdogrul O., and Erbilir F, Isolation and Characterization of Lactobacillus bulgaricus and Lactobacillus casei from various foods. Turk J Bio. 30:39-44 (2006).
- Forouhandeh H., Vahed Z.S., Hejazi M.S., Nahaei M.R., and Dibavar M.K., Isolation and Phenotypic Characterization of Lactobacillus species from various dairy products. Current Research in Bacteriology 3(2):84-88 (2010).
- Guessas B., and Kihal M., Characterization of lactic acid bacteria isolated from Algerian arid zone raw goats’ milk. African Journal of Biotechnology 3(6), 339-342 (2004).
- Mehmood T., Masud T., Abbass S.A., and Maqsud S., Isolation and Identification of wild strains of lactic acid bacteria for yoghurt preparation from indigenous dahi. Pakistan Journal of Nutrition 8(6):866-871(2009).
- Nair P.S., Surendran P.K., Biochemical characterization of lactic acid bacteria isolated from fish and prawn. Journal of Culture Collection (4), 48-52 (2005).
- Sameen , Anjum F.M., Huma N., and Khan M.I Comparison of locally isolated culture from yoghurt (dahi) with commercial culture for the production of mozzarella cheese. International Journal of Agricultural and Biology 12:231-236 (2010).
- Tserovska L., Stefanova S., and Yordanova T., Identification of lactic acid bacteria isolated from Katyk, Goat’s milk, Cheese. Journal of Culture Collection (3), 48-52 (2002).
- Viljanen M.J., Jääskeläinen S.A., Messner P., Sleytr U.B., and Palva A., Isolation of three new surface layer proteins genes (slp) from Lactobacillus brevis ATCC 14869 and characterization of the changes in their expression under aerated and anaerobic conditions. Journal of Bacteriology 184:6786-6795 (2002).

This work is licensed under a Creative Commons Attribution 4.0 International License.





