Manuscript accepted on : 18-Sep-2018
Published online on: 27-09-2018
Plagiarism Check: Yes
Mahdiyeh Pashaei1, Jamal EivaziZiaei2, Alireza Nikanfar3, Babak Emamalizadeh4 and Seyyed Mojtaba Mohaddes Ardebili5
1Hematology and Medical Oncology, Hematology and Oncology Research Center, Tabriz University of Medical Sciences, Department of Biochemistry and Clinical Laboratories, Division of Medical Genetics,Tabriz University of Medical Sciences, Tabriz, Iran.
2Hematology and Medical Oncology, Hematology and Oncology Research Center, Tabriz University of Medical Sciences, Iran.
3Tabriz University Hematology and Oncology Research Center, Shahid Ghazi Hematology and Oncology Ward.
4Department of Medical Genetics, Tabriz University of Medical Sciences, Tabriz, Medical Genetics.
5Department of Biochemistry and Clinical Laboratories, Division of Medical Genetics, Faculty of Medicine, Tabriz University of Medical Sciences, Tabriz, Iran.
Corresponding Author E-mail: mojtabamohaddesardebili197@gmail.com
DOI : http://dx.doi.org/10.13005/bbra/2675
ABSTRACT: Breast cancer is one of the main factors in the mortality of Iranian women. A large rearrangement genome is observed in most genes, especially in BRCA1 / BRCA2 genes lacking small mutations in breast cancer. Therefore, methods are needed to detect one or more exon deletions or their duplication. Therefore, the aim of this study was to determine the change in the number of copies of ATM, BRCA1, CHEK2, PTEN, and P53 genes in women with breast cancer in the East Azarbaijan region by MLPA method. This research is a descriptive study that was conducted randomly among 150 Azeri women with breast cancer who were referred to Tabriz Nour Najat Hospital; sixteen healthy people were selected as control samples. Deletion and duplication of ATM, BRCA1, P53, CHEK2 and PTEN genes were investigated using the MLPA method. The results showed that there was no pathogenicity mutation in these five genes. Therefore, it can be said that a large rearrangement genome in the East Azarbaijan province is very unlikely to lead to breast cancer in the area.
KEYWORDS: ATM; BRCA1; Breast Cancer; CHEK2; MLPA; P53 PTEN;
Download this article as:| Copy the following to cite this article: Pashaei M, EivaziZiaei J, Nikanfar A, Emamalizadeh B, Ardebili S. M. M. Study of Some Genetic Variants for Cancer in Women with Breast Cancer in the East Azarbaijan Region by MLPA Method. Biosci Biotech Res Asia 2018;15(3). |
| Copy the following to cite this URL: Pashaei M, EivaziZiaei J, Nikanfar A, Emamalizadeh B, Ardebili S. M. M. Study of Some Genetic Variants for Cancer in Women with Breast Cancer in the East Azarbaijan Region by MLPA Method. Biosci Biotech Res Asia 2018;15(3). Available from: https://www.biotech-asia.org/?p=31204 |
Introduction
Breast Cancer is the most common cancer in women and causes the highest mortality rates worldwide. The deaths from this cancer include 23% of all deaths from cancer. Studies in Iran show that this cancer is the most common cancer among women. one of the most important symptoms of breast cancer is the presence of a rounded, elliptic, tight mass, or painless in the chest, as well as enlargement of the armpit lymph nodes. Old age, menstruation at a low age, first pregnancy at high or never have children, mother or sister with breast cancer, Breast/chest radiotherapy, the mammary tissue that appears dense in the mammogram, get hormones such as estrogen and progesterone, drinking alcohol, whitening, smoking, diabetes and obesity are the main risk factors for breast cancer. Generally, breast cancer treatments are generally divided into systemic groups (chemotherapy, hormone therapy, etc.); local therapies (such as surgery and radiotherapy); surgery is the most important type of breast cancer treatment.
There are several factors involved in the incidence of breast cancer, most of which are sporadic, but part of the disease is due to the inheritance of certain mutant genes which inherited. The results of two important studies indicate that hereditary factors may account for 20-30% of breast cancer. In addition to the BRIPI, CHEK2, TGFB, TP53, PTEN BRCA2, and BRCA1 genes, a number of other genes have also been associated with an increased risk of breast cancer. Therefore, mutations in the genes CDH1, STK11, LKB1, BRCA1, BRCA2, P53, PTEN lead to familial breast cancer, which are highly penetrant genes Some of the other genes, ATM, CHEK2, BRIP1, PALB2, are called low-penetrance, which makes BC hereditary. Among these, the BRCA1 gene is located on the long arm of chromosome 17 (17q21). This gene has 24 exons and encodes a protein of the same name with the length of 1863 amino acids. BRCA1 and BRCA2 genes play an important role in DNA repair by recombinant hemolysis, maintaining chromosome stability, activating vulnerable DNA control points, and regulating the cell cycle. The P53 gene is known to be the most important tumor suppressor and is located on chromosome 17. The P53 gene codes for 53 kDa (393 amino acids) nucleophile phosphodiesterase. Studies have shown that P53 gene mutations are found to be 20-40% of breast cancer. Various stimuli, such as ionizing radiation, oxygen deficiency, nitric acid, chemotherapy and DNA damage, activate P53. In response to various stimuli, P53 undergoes various changes, and this activation can have many effects. The ATM gene encodes on chromosomes 11 (11-22 q22) and codes for a 350-kD serine/threonine protein kinase, which plays an important role in cellular response to Double-strand breaks (DSB) and maintains genomic stability.
CHEK2 is located on the chromosome 22q12.1, codes for 60KDa protein and contains 546 amino acids. CHEK2 is a serine/threonine kinase nucleus and has two subtypes CHEK1 and CHEK2. It plays an important role in delaying the cell cycle in normal and damaged cells, and in the three cell cycles G1, G2, and S have performance. CHEK2, which is activated as an intermediary responsible for transmitting signals from the ATM (which this activation is done by Thr68 phosphorylation). Also, the CHEK2 activated by the ATM, transmits the damage signal across the core. It also causes phosphorylation of the effective downstream proteins. The effect of CHEK2 is on the classical cell cycle, the Cdc25A, Cdc25C, P53, BRCA1 phosphorylation, the effect on the E2F transcription factor, DNA repair, apoptosis, and also determines the fate of the cell in the cellular damage response pathway. CHEK2 prevents damaged DNA replication and also an accumulation of harmful mutations in the cell population.
In this study, a large rearrangement genomic was investigated in P53, BRCA1, CHEK2, PTEN, and ATM genes in women with breast cancer in East Azarbaijan province using the MLPA (Multiplex ligation-dependent probe amplification) method.
Methods and Materials
Type of Study
This research is a descriptive study carried out on blood samples from 150 women with breast cancer in the population of the East Azarbaijan province.
Society, Statistical Sample and Sampling Method
In this study, among 150 patients 3 cases have bilateral breast cancer and 2 cases of breast and ovarian cancer and 145 non-hereditary breast cancer, 16 controls have been used and the age range used in this study was between the ages of 39 and 60.
Preparation of Blood Samples
Patients with breast cancer who were referred to Nour Nejat Hospital were first received written consent, and blood samples were poured into tubes containing EDTA. The blood samples were taken to the Genetics Department of Tabriz University of Medical Sciences. Then DNA extraction was performed using the salting-out method. Nanodrop was then used for quantitative DNA testing and samples larger than 50 ng were selected. During the extraction period, the samples were kept at -20 ° C. PCR DNA was used in order to quantitatively measure the DNA extracted, then the product was thrown onto the agarose gel and the Sharp bands were obtained.
MLPA
In this study, the SALSA MLPA Kit P190-C1 CHEK2 (MRC-Holland) kit was used to perform the MLPA reaction. There are certain probes for the CHEK 1100del C mutation. There are also two probes right above CHEL2 in the 22q12 area. There are also 7 diagnostic probes for the ATM gene and 5 probes for the PTEN gene and a probe for the KLLN gene on both sides. There are two TP53 probes and one probe for the BRCA1 promoter. Finally, 8 of these main probes in Probemix detect several different positions in the autosomal chromosomes, and the length of the reproduction products is 132 and 481 bp.
The Q-components are not ligation-dependent, and in small quantities, if the MLPA reaction is successful, the size of the Q-sections is reduced. As the amount of DNA increases, the pattern of the curve of the Q pieces is shortened. On the other hand, D is just like other probes, that are DNA dependent and ligated. These pieces only appear when the ligation is done. The reactions were performed according to the instructions of the kit. Subsequently, samples were analyzed by ABI 131 Capillary Electrophoresis.
Results
Results of DNA Extraction from Samples
DNA samples were extracted by a saturated salt method from the peripheral blood. The OD nanotrape was then taken and the concentration of the samples was diluted to conduct a MLPA reaction in the range of 50-250 ng. The PCR was then placed on the gel using primers in the laboratory of the PCR genetic group. Electrophoresis gel Photocell of DNA samples (Fig. 1) showed that extracellular DNA quality was good.
 |
Figure 1: PCR results.
|
MLPA Results for Patient and Control Samples for P53, BRCA1, CHEK2, ATM, PTEN Genes
Based on the results of probes used for P53 gene in this study, no large genomic regeneration was observed for this gene. Also, the results of the examination of the probes studied for the CHEK2 gene showed that in this study 16 probes were present for this gene. and some probes have the Revers state, but no mutation has been observed for the gene and the results for this gene were normal (fig.2)
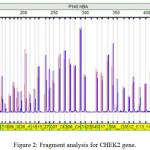 |
Figure 2: Fragment analysis for CHEK2 gene.
|
In Fig. 2, the size of the blue reference peaks are identical with the sample peaks that are red, indicating that the target person is normal.
The results also showed that the Peaks size is as a reference sample, and the peak ratio is equal to one, indicating no elimination or addition (normalization) of these genes in the patient (Fig. 3).
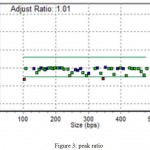 |
Figure 3: peak ratio.
|
Examined probes for the ATM gene showed no specific mutations for this gene, but there was an obscure mutation in this gene, but because of the low quality of DNA, it cannot be said which exon there is deletion (Fig. 5)
 |
Figure 4: Possibility of mutation in the ATM gene.
|
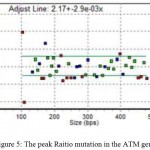 |
Figure 5: The peak Raitio mutation in the ATM gene.
|
A review of PTEN gene probes showed that no specific mutation was found for this gene. Also, the results of the probe and exon analysis of the KLLN gene showed that this gene is located on chromosome 10 and is regulated by P53, and there is also a probe for this gene, but no deletion and addition.
Studies have shown that there is only one probe for the BRCA1 gene and it’s in Exon 1, 2, and 13 and there is no mutation in this gene, it can be due to the lack of proper probes. So, in summary, we can say that the results from Fragment analysis of the genes of the marker V.G.2.6.0. showed that the patient samples, like the control samples, lacked any deletion or addition. The result obtained from this study shows that LGR in the P53, ATM, PTEN, CHEK2, BRCA1 genes does not cause breast cancer in these 150 patients.
Discussion and Conclusion
Breast cancer, which is a heterozygote and hormonal disease, which is 22.9% and the second most common cancer among women in the most popular countries that of all 8 women, one woman has this condition, but in every 1,000 men, one person suffered from this disease during his life. Breast cancer is a complex heterogeneous disease that involves many risk factors including geographical variables, lifestyle, first gestational age, menstrual period, menopause age and familial history.
Molecular studies show that those who inherit mutations in the BRCA1 / BRCA2 genes during their lifetime show a higher risk of developing the disease by 80-38% than the general population of the Americas with a risk of 13.5%. According to studies conducted in Iran, BC patients are one decade younger than in other countries, indicating that most cancers in Iran are inherited. According to new studies that show that breast cancer in developing countries is increasing, mutations in the ATM, BRCA1/BRCA2, CHEK2, MSH2, MSH6, PTEN,P53, STK1, BARD1, MUTYH, BRIP1, CDH1, MLH1, MRE11, NBN, PALB2, PMS1, PMS2, RAD50, are responsible for increasing the risk of breast cancer in women. Rare mutations cause 2 to 5% of breast cancer. However, mutations in reproductive cells of some of the genes that are highly influential genes include BRCA1, BRCA2, PTEN, ATM, P53, CHEK2 PPM1D genes which are commonly found in the incidence of breast cancer, mutations in these genes in reproductive cells contain 25% of familial breast cancer. In this study, all of the person examined for the rearrangement genome for the ATM, CHEK2, BRCA1, PTEN, and P53 were normal, without any deletion and duplication. In this study, there is only one probe available for the BRCA1 gene, for the promoter of this gene that in any of the studies we examined, there was no deletion in the promoter, and in addition, the LGR, according to studies, is a relatively rare process, and the prevalence of LGR in Asian countries is very low. And in this study, which 150 patients were normal, it can be concluded that the occurrence of LGR in East Azarbaijan province is a rare process and can not be the cause of breast cancer. Also, another study that was conducted in Iran and collected patients from different parts of the country, there was no elimination or duplication in BRCA1 / BRCA2 genes in this study.
According to researchers Lerman, Seag, Balshem, and Audrian, healthy relatives of Ovarian or breast cancer have a tendency to exaggerate the level of risk they are posing. More recently, genetic tests have been conducted to provide information to healthy women in families at risk for lifestyle changes and early detection of breast cancer.
Manguoğlu et al. reviewed the CHEK2, BRCA1, BRCA2 genes in Turkey in 2011, and did not find LGR for any of the genes, and they suggested that a large rearrangement genomic in these genes in Turkey is very low.
Aydin et al. (2011) conducted a study in Turkey that examined the BRCA1 / BRCA2 gene, which in the BRCA1 gene, exons 21, 18, and 19 were deleted in three patients but lacking any deletion or duplication in the BRCA2 gene.
Also, a study by Silva and colleagues in 2012 showed that they could not find mutations in BRCA2 and P53 genes, and found that the BRCA1 gene found a mutation that had a deletion between intron 8 and intron 19 in one patient and they identified the cause of deletion in this gene is the sequence of ALU.
A study of the Sri Lankan population by Tennekoon et al. in 2014 has no deletion in the BRCA1 / BRCA2 gene. They stated that LGR is very low in Asian countries, and LGR is also Founder effect in some regions.
Tedaldi et al. In 2014 conducted the study in Helsinki. They found only one duplication of the patient at CHEK2 at Exon 6-13 and in two healthy individuals. The disease patients they examined had hereditary breast cancer. They checked that this duplication was not due to the pseudogenes sequence in the gene. And this duplication triggers a frameshift in codon 488 that involves the second kinase of the product of this gene, It also plays a role in proteins involved in repairing DNA damage, which leads to the creation of codon 503 and early termination. However, in none of the BRCA1 / BRCA2 genes couldn’t found the mutation. They also stated that the gene was inherited as low penetrance and mutation, which would very likely cause breast cancer in women and prostate cancer in men. They claimed that this mutation is rare but can significantly contribute to cancer development. These findings suggest that methods that are commonly used to search for mutations are defective in determining the exact type of BRCA1 defects in reproductive cells, such as large Intergenic invertebrates.
The loss or full extension of an exon or a proliferation in the case of a wild type allele may result in false negative results. Because a wild type allele is followed by a positive PCR. Additionally, the large recombination creates FrameShift, which results in an early end in transcription/translation. Therefore, the patient phenotype with this mutation type cannot be distinguished from patients with other mutations. Large deletions will one or more exons, while they can cause loss of important areas of operation BRCA1 (Functional Domain).
A powerful method for investigating and identifying major LGR rearrangement genomic changes in a large number of samples is the MLPA method. This method is much simpler than the previous methods and makes larger values on the gene structure and minimizes the false negative ones.
Considering that BRCA1 / BRCA2 genes are two of the most important genes in hereditary breast cancer, these two genes make up 15-20% of BC’s heredity, while 5 to 10% of the total BC is hereditary. Since most of the mutations in these two genes are small mutations, however, large LGR mutations have also been seen in different ethnic groups.
Using the SALSA P002 / P002B kit BRCA1 (MRC-Holland Amsterdam, Holland), a study done in Italy on 221 patients. The result showed that among three patients, the exons 11-13 were eliminated in one, exons 13-15 were eliminated in the other and in the last one, all 24 exons were eliminated.
Conclusion
In general, the study found that in the population studied, there were five common genes that caused mutations in the reproductive cells of individuals in the ATM, BRCA1, P53, CHEK2, and pten genes, which led to an inherited breast cancer, leading to an early age-related cancer. In these five studied genes, there is no elimination or duplication. Thus, LGR cannot lead to breast cancer in ATM, PTEN, P53, CHEK2 genes. In the case of the BRCA1 gene, which in most other studies has deletion or duplication, but in this study only has a probe on this gene, which is also related to the promoter, which in most other studies mostly involves exons 12 and 13.It is also due to the ALU sequence and the Pseudogene of the 5 genes and the unequal recombination of this gene.
The MLPA method can justify the genetic factor of the disease in some cases without specific factors and determine their genetic defect. By identifying this defect in a family, it can be used to manage the disease in other people at risk for the family.
Therefore, the MLPA technique should be used as a diagnostic method, along with common molecular techniques, to diagnose people at risk for breast cancer in order to increase the ability to identify those who carry the mutation that causes breast cancer and ultimately causes improve management and control of these patients in the community.
References
- Khodarahmi M., Azadbakht L. The Association between Different Kinds of Fat Intake and Breast Cancer Risk in Women. Int J Prev Med. 2014;5(1):6.
- Mousavi S. M., Mohaghegi M. A., Mousavi-Jerrahi A., Nahvijou A., Sseddighi Z. Outcome of breast cancer in Iran a study of Tehran Cancer Registry data. Asian Pac J Cancer Prev. 2008;9(2):275-8.
- Falagas M., Zarkadoulia E., Ioannidou E., Peppas G., Christodpulou C. h., Rafailidis P . The effect of psycho social factors on breast cancer outcome a systematic review. Breast Cancer Research. 2007;9:234-244.
CrossRef - Namiranian N., Moradi-Lakeh M., Razavi-Ratki S. K., Doayie M., Nojomi M. Risk factors of breast cancer in the Eastern Mediterranean Region: a systematic review and meta-analysis. Asian Pac J Cancer Prev. 2014;15(21):9535-41.
CrossRef - Silva F., Romero V., Carvalho A. L., Borgo M., Amorim M. H., Gouvea S., et al. Hormone therapy with tamoxifen reduces plasma levels of NT-B-type natriuretic peptide but does not change ventricular ejection fraction after chemotherapy in women with breast cancer. Brazilian Journal of Medical and Biological Research. 2014;1414-431.
CrossRef - Al-Moghrabi N., Nofel A., Al-Yousef N., Madkhali S. M., Amer B. S., Alaiya A., et al. The molecular significance of methylated BRCA1 promoter in white blood cells of cancer-free. BMC Cancer. 2014;14:830.
CrossRef - Zhang J., Willers H., Feng Z., Jagadish C., Kim S., David T. Chk 2 Phosphorylation of BRCA 1 Regulates DNA Double-Strand Break Repair. Mol Cell Biol. 2004;24(2):708–718.
CrossRef - Chabalier-Taste C., Racca C., Dozier C. H., Larminat F. BRCA 1 is regulated by Chk 2 in response to spindle damage. Biochimica et Biophysica Acta (BBA)Molecular Cell Research. 2008;(12):2223–2233.
- Foroughizadeh M., Mozdarani H., Majidzadeh A. K., Kaviani A. Variation of ATM Gene Expression in Peripheral Blood Cells of Sporadic Breast Carcinomas in Iranian Patients. Avicenna J Med Biotechnol. 2012;4(2):95–101.
- Calvez-Kelm F., Lesueur F., Damiola F., Vallée M., Voegele C., Babikyan D., Durand G.,et al. Rare evolutionarily unlikely missense substitutions in CHEK2 contribute to breast cancer susceptibility results from a breast cancer family registry case-control mutation-screening study. Breast Cancer Res. 2011;13(1):6.
CrossRef - Schmidt M., Tollenaar R., Kemp S., Broeks A., Cornelisse C., Smit V.,et. Breast Cancer Survival and Tumor Characteristics in Premenopausal Women Carrying the CHEK2*1100delC Germline Mutation. Journal of Clinical Oncology. 2006;25(1)64-69.
CrossRef - Craig A., Hupp T. The regulation of CHK 2 in human cancer. Oncogene. 2004;23:8411–8418.
CrossRef - Kullmann K., Deryal M., Ong M. F., Schmidt W., Mahlknecht U. DNMT 1 genetic polymorphisms affect breast cancer risk in the central European Caucasian population. Clinical epigenetics. 2013;5(1):7.
CrossRef - Calvez-Kelm F., Lesueur F., Damiola F., Vallée M., Voegele C., Babikyan D., Durand G.,et al. Rare evolutionarily unlikely missense substitutions in CHEK2 contribute to breast cancer susceptibility results from a breast cancer family registry case-control mutation-screening study. Breast Cancer Res. 2011;13(1):6.
CrossRef - V. R. The methylenetetrahydrofolate reductase C677 T polymorphism and breast cancer risk in asian populations. Asian Pac J Cancer Prev. 2014;15.
- Panchal S. M., Ennis M., Canon S.,Bordeleau L. J. Selecting a BRCA risk assessment model for use in a familial cancer clinic. BMC Medical Genetics. 2008;9:116.
CrossRef - Foroughizadeh M., Mozdarani H., Majidzadeh A. K., Kaviani A. Variation of ATM Gene Expression in Peripheral Blood Cells of Sporadic Breast Carcinomas in Iranian Patients. Avicenna J Med Biotechnol. 2012;4(2):95–101.
- Aydin F., Akagun T., Yildiz B., Fidan E., Ozdemir F., Kavgaci H. Clinicopathologic characteristics and BRCA-1/BRCA-2 mutations of Turkish patients with breast cancer. Bratisl Lek Listy. 2011;112(9):521-3.
- Silva A. G., Ewald I., Sapienza M., Peixoto P., Nóbrega A.,et al. Li-Fraumeni-like syndrome associated with a large BRCA1 intragenic deletion. BMC Cancer. 2012;12:237.
CrossRef - Aretz S., Stienen D., Fried W. High proportion of large genomic deletions and a genotype–phenotype update in 80 unrelated families with juvenile polypos is syndrome. J Med Genet. 2007;44(11):702-709.
CrossRef - Tedaldi G., Danesi R., Zampiga V.,Tebaldi M., Bedei L.,Zoli W.,et al. First evidence of a large CHEK2 duplication involved in cancer predisposition in an Italian family with hereditary breast cancer. BMC Cancer. 2014;14:478.
CrossRef - Aldred M. A., Vijayakrishnan J., James V., Soubrier F., Gomez-Sanchez M. A., Martensson G., et al. BMPR 2 gene rearrangements account for a significant proportion of mutations in familial and idiopathic pulmonary arterial hypertension. Hum Mutat. 2006;27(2):212-3.
CrossRef - Aretz S., Stienen D., Fried W. High proportion of large genomic deletions and a genotype–phenotype update in 80 unrelated families with juvenile polyposis syndrome. J Med Genet. 2007;44(11):702-709.
CrossRef - Beetz C., Nygren A. O. H., Schicke J., Auer-Grumbach M., Bürk K., Heide G., et al. High frequency of partial SPAST deletions in autosomal dominant hereditary spastic paraplegia. Neurology. 2006;67(11):1926-30.
CrossRef - Silva S., Tennekoon K. H., Karunanayake E. H., Amarasinghe I., Angunawela P. Analysis of BRCA1and BRCA2 large genomic rearrangements in Sri Lankan familial breast cancer patients and at risk individuals. BMC Res Notes. 2014;7:344.
CrossRef

This work is licensed under a Creative Commons Attribution 4.0 International License.





