Manuscript accepted on : 29 March 2018
Published online on: --
Plagiarism Check: Yes
Cultural, Anastomosis, and Universally Primed PCR Typing of Rhizoctonia Solani from Potato
Department of Biology, Science and Humanities College, Alquwayiyah, Shaqra University, Saudi Arabia.
Corresponding Author E-mail: malghuthaymi@su.edu.sa
DOI : http://dx.doi.org/10.13005/bbra/2614
ABSTRACT: Forty two isolates of Rhizoctonia solani were recovered from potato in four agro-ecological regions of potato in Saudi Arabia were studied by using anastomosis typing, morphological characters, and molecular method. Almost of isolates sampled from potato cultures belonged to AG-1, AG-3, and AG-7. RP1, RP2, RP3, and RP4 are bridging isolates, they might be classified into another anastomosis groups (AGs) or perhaps might be new anastomosis groups. Universally primed PCR (UP-PCR) fingerprinting was used to evaluate genetic diversity of R. solani AGs infecting potato and other hosts. The majority of the isolates representing various AGs were grouped together into different sub-clusters using all three primers. Molecular groups of the isolates did not correspond to agro-ecological regions or states and crops of the origin. In the current study, the UP-PCR markers could not identify and clearly differentiate the isolates of R. solani. There was no relationship between the geographic origin of the isolates and clustering of isolates based on the genetic diversity. Based on cultural characterization, anastomosis group typing and UP-PCR, no clear-cut conclusions about non-identified anastomosis groups from potato.
KEYWORDS: Anastomosis Typing; Potato; Thanatephorus Cucumeris; Sclerotia; UP-PCR
Download this article as:| Copy the following to cite this article: Alghuthaymi M. A. Cultural, Anastomosis, and Universally Primed PCR Typing of Rhizoctonia Solani from Potato. Biosci Biotech Res Asia 2018;15(1). |
| Copy the following to cite this URL: Alghuthaymi M. A. Cultural, Anastomosis, and Universally Primed PCR Typing of Rhizoctonia Solani from Potato. Biosci Biotech Res Asia 2018;15(1). Available from: https://www.biotech-asia.org/?p=29552 |
Introduction
Currently, Saudi Arabia is self-sufficient in potato production, with an area of 17665 ha (KACST, 2012). In 2013, the main potato producing areas in Saudi Arabia were Al-Jouf, Hail, Tabouk, Hofof, Qasim, Riyadh, and Najaran, with a total production area of 17,500 ha and total production of 460,000 tons per year of which 45,000 tons were seed potato (FAOSTAT, 2015). Rhizoctonia solani Kühn (teleomorph = Thanatephorus cucumeris (Frank) Donk) the most widely recognised species of Rhizoctonia, was described on potato in 1858 (Ogoshi, 1996). R. solani is the causal organism of Rhizoctonia disease complex in potato (Wilson et al. 2008) resulting in two different appearances of the disease, namely stem canker and black scurf. R. solani anastomosis group AG- 3 is a common fungal inhabitant of the Potato tuber rhizosphere and has a universal distribution (Cubeta and Vilgalys, 2000, Fiers et al. 2011). Recently, R. solani has developed into an important pathogen in potato growing-areas in Saudi Arabia (El-Hussieni, 2004, Abd-Elsalam et al. 2009a,b). R. solani isolates differ in phenotypic and genotypic characteristics, and have been traditionally arranged in genetically related groups based on hyphal anastomosis grouping. Currently, 14 AGs have been identified (AG-1 to AG-13 and AG-BI) on different plant species (Carling et al. 2002). The classical diagnostic methods of fungal pathogens are time consuming and sometimes difficult to interpret. For instance, determination of AGs by hyphal anastomosis needs to microscopic experience, and it is a time-consuming method. Also, differentiation of R. solani isolates based on phenotypic characters has proved be very tedious and less reliable. PCR-based markers can be used for study genetic diversity in Rhizoctonia has resulted in an improved understanding of the basis of habitat and host-specificity of member species and subgroups of R. solani. Such markers have provided useful tools for differentiating isolates within an anastomosis group (AG) and those belonging to diverse AGs (Abd-Elsalam et al. 2009). Numerous genetic markers were developed to evaluate genetic variability and characterization of R. solani such as, Universal Rice Primers (URP) (Pathania et al. 2010, Mishra et al. 2015), Random amplified polymorphic DNA (RAPD) (Banerjee et al. 2012) whereas inter-simple-sequence repeat (ISSR) is a simple, cost efficient, robust, multilocus marker method for determining genetic variability among the isolates of pathogens (Yugander et al. 2015). Universal-Primed PCR (UP-PCR) could discriminate AGs of R. solani by cross-blot hybridization. They identified 16 isolates of R. solani into their AG subgroup by using UP-PCR method (Lûbeck and Poulsen, 2001). In particular, UP-PCR hybridization can be a useful procedure to fast screen and link a large number of unidentified Rhizoctonia isolates to their AGs and AG subgroups. The sequence data of unique UP-PCR fragments can be used to design specific primers for detection of R. solani AG 1-IB and AG 2-2IIIB (Amaradasa et al. 2014). Little is known about the predominant AG associated with potato diseases in different geographic regions Saudi Arabia. The main aim of this study was to determine which AGs are associated with potato crops producing-areas in Saudi Arabia by morphological characterization, observations of hyphal interaction and molecular characterization using UP-PCR technique.
Materials and Methods
Collection and Isolation of R. Solani Isolates
Potato plants and tubers showing typical R. solani symptoms (black scurf, elephant hide, and canker on stems, stolons and roots) and atypical symptoms (malformation, trumpet holes and cavities on growth cracks) together with potato rhizosphere soils were sampled from four potato growing regions (Wadi Al-Dawasir, Kharj, Hail, and Qasim) in Saudi Arabia during 2016. The fungal pathogens were isolated and purified following the standard protocol (hyphal tip culture method) using water agar and potato dextrose agar media (Abd-Elsalam et al. 2009a). The isolates were maintained in PDA 2-ml micro tubes at 4°C.
Cultural Variability, Nuclei Number and Anastomosis Grouping
R. solani isolates were transferred on three types of media including; potato malt agar (PMA) (Himedia, India), malt yeast extract (MYA), and potato dextrose agar (PDA) in Petri dishes to study in detail for their cultural and morphological characteristics. The observations on colony colour and aerial mycelium texture were recorded at 7 days after incubation and sclerotial characters of each isolate observed after 15 days. Three replications were kept for each isolate. R. solani isolates were identified morphologically by examining the hyphal branching, and the septal pore type. Twenty randomly selected cells per isolate were assayed, and the number of nuclei per cell was counted. Before assay, hyphae were stained with a DAPI technique in the vegetative cells (Kulik and Dery, 1995) and the hyphae were stained with a safranin-O (0.03%) and KOH (3%) solution and then observed under brightfield microscopy at 400 magnification. Anastomosis grouping (AG) can be a key indicator of somatic and vegetative compatibility between isolates and was conducted by pairing unknown isolates with known tester strains. For typing of anastomosis group, collected isolates from Saudi Arabia and 10 tester strains were paired by plating on unknown isolate opposite a known AG type on 2% water agar (WA) in Petri dishes. The fungal cultures were incubated under cool white light with 12 h photoperiod at 25°C. Observation of hyphal fusion was carried by observing hyphal interaction zone at 40 x magnification using light microscope. Hyphal interactions were assigned to one of the four categories according to MacNish et al. (1997), i.e. C0 = no recognition observed between hyphae, C1 = hyphal contact, connection of walls but no membrane to membrane contact, C2 = hyphal fusion resulting in the death of fused and adjacent cells, and C3 = fusion of walls and membrane and no evidence of cell death. Anastomosis was regarded as positive when hyphae between the potatoes isolate and AG tester made contact with each other and their walls fused, with subsequent plasmolysis of adjacent cells. R. solani tester isolates (AG-1, AG-3; AG-4 AG-5; AG-7; AG-8; AG-9; AG-10; AG-11; AG-12; and AG-13) was obtained from Plant Pathology Research Institute, Agricultural Research Centre, Giza, Egypt.
DNA Extraction
Only 14 isolates used in the molecular characterization was given in Table 1. R. solani isolates were grown in potato dextrose broth (PDB) medium at 28°C for at least 5 days. The mycelia were harvested by vacuum filtration, then ground to a fine powder with liquid nitrogen. Rhizoctonia mycelium (100 mg) were homogenized in 400 µl of sterile salt homogenizing buffer (200 mM Tris-HCl, pH 8.5, 250 mM NaCl, 25 mM EDTA, 0.5% SDS). Then, 6 µl of 20 mg ml–1 RNase A (20 mg ml–1 final concentration) were added and mixed well. The micro tubes were incubated at 65°C for 10 min, after which 130 µl of 3 M sodium acetate, pH 5.2 was added to each sample. Samples were vortexed for 30 s at maximum speed, and incubated at –20°C for 10 min. The lysate was centrifuged at 12000 rpm at 4°C for 15 min. The supernatant was transferred to new tubes. An equal volume of isopropanol was added to each sample, mixed well, and samples were incubated at –20°C for 10 min. Samples were then centrifuged for 20 min at 4°C, at 7000 rpm. Washing the DNA pellets was performed twice using 700 µl of washing solution (100% and 70% ethanol, respectively). The DNA pellets were subsequently air dried in an oven at 40°C for at least 10 min. The DNA was dissolved in 100 µl of sterile distilled and deionized water, and its quality was checked through electrophoresis.
UP-PCR Conditions
For molecular characterization one isolate for each new R. solani groups was selected based on their similar in cultural, microscopic and anastomosis typing (RP1, RP2, RP3 and RP4) in additions 10 R. solani testers as a references isolates. Amplification reactions were performed in 0.2- ml micro centrifuge tubes in a 25-µl reaction volume containing 10 mM Tris-HCl, pH 8.8, 50 mM KCl, 0.8 mM NaCl, 3.5 mM MgCl2, 0.1% Triton X- 100, 0.4 mM dNTPs, 40 ng primer L45 (5‘-GTAAAACGACGGCCAGT-3‘), L21 (5‘ GGATCCGAGGGTGGCGGTTCT-3‘), and AS15 (5-GGCTAAGCGGTCGTTAC-3‘)1.0 U of Taq DNA polymerase (Promega, Mannheim, Germany), and 10 to 15 ng of genomic DNA. PCR amplification was performed in a Biometra Thermal Cycler System (Germany) programmed for 30 cycles of denaturation at 94°C for 30 s (first denaturation step at 94°C for 3 min), annealing at 56°C for 70 s and polymerization at 72°C for 60 s, with a final extension step of 72°C for 5 min. The reaction tubes were held at 4°C following the final amplification cycle. Electrophoresis of PCR-amplified products was performed in 1.5% agarose gels for 1.5 h at 7.0 V/cm2. PCR products were stained with 0.5 g/ml of ethidium bromide and visualized with 305 nm ultraviolet light. UVIsoft analysis (Gel Documentation and Analysis Systems, Uvitec, Cambridge, UK) was used to capture the images and to calculate unweighted pair group method with arithmetic mean (UPGMA) cluster.
Hierarchical Clustering
Fungal culture-associated parameters of R. solani isolates were compared based on dendogram illustration. A hierarchical clustering analysis was performed and dendrogram was constructed by SPSS Statistics software (Version 20).
Results
Fungal Morphology and Anastomosis Grouping
In the current study forty two R. solani isolates were collected as following; 9 isolates from Wadi Al-Dawasir, 16 isolates from Kharj, 8 isolates from Hail, and 9 isolates from El-Qasim) (Fig. 1). All collected isolates was multi-nucleate, forming 3 to 14 nuclei per cell, providing further support for the isolate being identified as belonging to the species R. solani. Based on their cultural properties on three different cultural media, of the 42 isolates analysed, 30 were isolated from stem cankers and 12 from sclerotia on mother tubers. Of the 42 isolates tested, 3 belonged to AG-1, 7 to AG-3, and 2 to AG-7, the remaining 12 isolates failed to anastomose with any of the testers used (Fig. 1). Several potential non-AG3 isolates were also tentatively identified in this process. In current study four new R. solani groups RP1 (7 isolates), RP2 (4 isolates), RP3 (5 isolates) and RP4 (2 isolates) were identified. They might be classified into another anastomosis group or perhaps might be new anastomosis groups (Fig. 1). The microscopic features associated with R. solani, such as hyphae with regular septa, right-angled branching pattern and multinucleate (Fig. 2). Therefore, we believe that RP1, RP2, RP3, and RP4 are bridging isolates, or they are special groups of R. solani in Saudi Arabia ecological regions (Fig. 3).
 |
Figure 1: Frequency and distrbution of R. solani isolates based on their geographic orgins and anastomosis groups (AGs)
|
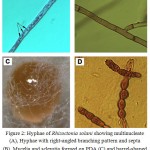 |
Figure 2: Hyphae of Rhizoctonia solani showing multinucleate (A), Hyphae with right-angled branching pattern and septa (B), Mycelia and sclerotia formed on PDA (C) and barrel-shaped moniliod cells in chain, 14 days at 28°C on PDA (D).
|
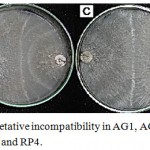 |
Figure 3: Anastomosis and vegetative incompatibility in AG1, AG 3, AG4, AG7 and tip subcultures for RP1, RP2, RP3 and RP4.
|
(A) Sparse tuft formed between AG3 and field isolate RP4 (B) Mild tuft formed between AG1 and field isolate RP2 (C) Compact tuft formed between AG4 and field isolate RP1 and RP4 and (D) Weak incompatibility formed between AG7 and field isolate RP3.
Cultural Characterization
Unidentified R. solani isolates (RP1) from potato-producing areas in Saudi Arabia had non aerial mycelia, without zonation, and cream main hyphae on three types of cultural media after 2 weeks of incubation. Isolates of R. solani (RP2) had aerial non-aerial mycelia, with and without zonation, and dark brown hyphae on three types of cultural media (Table 2). RP4 isolates were buff in color early in the growth development stage but turned to a dark brown color within two weeks on PDA and MYA media, also had regular clusters of mycelia (sclerotia), zonation or concentric rings, and zonation on all types media (Fig. 4).
 |
Figure 4: Colony appearance, Mycelial and sclerotial growth of R. solani AGs s on three different types of media (PDA, MYA, and MPA).
|
Hierarchical clustering analyses between 14 examined R. solani isolates which are clustered based on culture-associated parameters including aerial mycelium, sclorotia and zonation on three different types of media (PDA, MYA, and MPA). The phylogenetic analysis based on culture-associated parameters revealed two major clusters of R. solani collected from Saudi Arabia and AGs testers, and one minor cluster was included AG-13 with 75% genetic similarity. First main cluster consisted of seven isolates while the six isolates were grouped into second main cluster. First main cluster contained AG-3, AG-5, AG-7, AG-9, AG-10 and one isolate of undetermined AGs (RP4) with 98% genetic similarity to AG-12. Second main cluster contained AG-1, AG-4, AG-8 and three isolate of undetermined AGs (RP1, RP2 and RP3). RP1 was closely related to AG-4 with 92% genetic similarity while, RP3 closely related to AG-1 and AG-8 with 92% genetic similarity. No correlation was observed between cluster analysis and fungal cultural parameters for different AGs of R. solani isolates (Fig. 5).
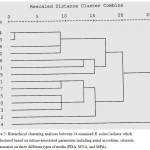 |
Figure 5: Hierarchical clustering analyses between 14 examined R. solani isolates which are clustered based on culture-associated parameters including aerial mycelium, sclorotia and zonation on three different types of media (PDA, MYA, and MPA).
|
UP-PCR
The primers L45, L21 and AS15 produced multi-bands PCR products of molecular weight ranging from 200 bp to 1100 bp in different isolates with diverse amplicon. To test the resolving ability of these primers, cluster analysis of the resulting data were performed using the Unweighted Pair-Group Method with Arithmetic Average method. The resulting dendrogram for L45 primer categorized the isolates into two groups, and one minor cluster was included AG-4 with 70% genetic similarity. Thirteen R. solani isolates were classified into 2 major groups, RP1 and AG-13, AG-7 and AG-10, RP4 and AG-3 (94% similarity) are closer related to each other than to other AGs. Also, RP3 and RP4 are closer related to AG-1 and AG-3 with a genetic similarity 78% (Fig. 6). As depicted in the phenogram, only small groups of isolates with identical pattern could be identified. Isolates were clustered into two major groups linked at 60 and 78% similarity. RP1, RP2 and RP4 are closer related to AG-4 and AG-7 with a genetic similarity 78%. The L21 primer region formed four groups without any clear cut differentiation among isolates and did not follow the pattern of UP-PCR analyses (Fig. 7). The UPGMA dendrogram demonstrated that the isolates of R. solani were grouped into two distinct clusters while the genetic similarity coefficient ranged from 62 to 72%. A good similarity was found between RP1, RP3 and AG-5 with a genetic similarity 78%, while RP4 showed the highest similarity with the AG-1 and AG-13 isolate at the genetic similarity of 85%. This analysis confirmed the wide variability observed within RP isolates and the testers used in the current study (Fig. 8). Not all of the primers used successfully with other R. solani subgroups amplified products or were informative for distinguishing polymorphism among the isolates used in this study. The majority of the isolates representing various AGs were grouped together into different sub-clusters using all three primers. Molecular groups of the isolates did not correspond to agro-ecological regions or states and crops of the origin. In the current study, the UP-PCR markers could not identify and clearly differentiate the isolates of R. solani.
 |
Figure 6: UPGMA dendrogram for Rhizoctonia isolates derived by universally primed PCR markers (primer L45) representing genetic relatedness among 10 anastomosis group testers, 4 isolates collected from potato.
|
The scale above the dendrogram represents genetic similarity coefficients calculated according to Pearson’s correlation coefficient.
 |
Figure 7: UPGMA dendrogram for Rhizoctonia isolates derived by universally primed PCR markers (primer L21) representing genetic relatedness among 10 anastomosis group testers, 4 isolates collected from potato.
|
The scale above the dendrogram represents genetic similarity coefficients calculated according to Pearson’s correlation coefficient.
 |
Figure 8: UPGMA dendrogram for Rhizoctonia isolates derived by universally primed PCR markers (primer AS15) representing genetic relatedness among 10 anastomosis group testers, 4 isolates collected from potato.
|
The scale above the dendrogram represents genetic similarity coefficients calculated according to Pearson’s correlation coefficient.
Discussion
The main objective of this study was to characterise isolates of R. solani from Saudi potato
plants using the following methods: hierarchical clustering patterns based on culture-associated parameters, anastomosis grouping, and universally primed PCR for molecular characterization of R. solani isolates. In the present study, the fungus was multi-nucleate, forming 3 to 14 nuclei per cell, providing further support for the isolate being identified as belonging to the species R. solani. Microscopically all Rhizoctonia species look similar in the asexual state and typically characterized by nonsporulating mycelia with 90 degree branches having dolipore septa (Smiley et al. 2005).
A system of anastomosis grouping based on hyphal fusion is widely accepted as the basis for recognizing groups within the species complex (Sneh et al. 1991). Our isolates from potato 3 belonged to AG-1, 7 to AG-3, and 2 to AG-7 anastomosis groups. Four new R. solani groups RP1 (7 isolates), RP2 (4 isolates), RP3 (5 isolates) and RP4 (2 isolates) were identified based on their cultural characterization, anastomosis typing and frequency. They might be classified into another anastomosis group or perhaps might be new anastomosis groups. Differing anastomosis reactions may be observed when genetically distinct hyphae from the one colony are opposed to those of a second colony, resulting in the incorrect classification of an isolate as a particular AG, or the inability to place an isolate into a specific AG subgroup, as seen in this study. Therefore, we believe that RP1, RP2, RP3, and RP4 are bridging isolates, or they are special groups of R. solani in Saudi Arabia ecological regions. Rhizoctonia disease in potato fields in Saudi Arabia is caused by a mixture of disparate anastomosis groups (Abd-Elsalam et al. 2009). No correlation was observed between cluster analysis and fungal cultural parameters for different AGs of R. solani isolates. However, most previous work concludes that AG3-PT (the potato subgroup) is the predominant AG in potato crops (Tsror 2010). AG3-PT is one of three subgroups of AG3, the other subgroups being AG3-TB, the tobacco subgroup, and a new subgroup consisting of tomato isolates (Bartz et al. 2010).
Since most investigations of this type have used the hyphal fusion method to determine AG, such difficulties can be understood. Also, morphological characters are also influenced by environmental and cultural conditions. To overcome such failures, a combination of molecular and hyphal fusion methods were used in more recent studies study to determine the relative incidence of anastomosis groups present in potato crops (Blazier and Conway, 2004, Woodhall et al. 2007). Universal primers (UP) are longer (15–21 nt) than conventional RAPD primers (typically 10 nt) and designed to anneal under more stringent conditions (52–60 C) ensuring higher reproducibility of banding profiles (Trigiano et al. 2004, Lûbeck and Lûbeck 2005). Lûbeck and Poulsen (2001) found considerable variation in UP-PCR banding profiles among isolates of R. solani from AGs and AG subgroups. The UP-PCR analysis can be used to overcome the variation observed in ITS-PCR because it tests multiple loci simultaneously.
Not all of the primers used successfully with other R. solani subgroups amplified products or were informative for distinguishing polymorphism among the isolates used in this study. The majority of the isolates representing various AGs were grouped together into different sub-clusters using all three primers. Molecular groups of the isolates did not correspond to agro-ecological regions or states and crops of the origin. In the current study, the UP-PCR markers differentiate R. solani isolates but did not give clear structuration.
Conclusion
Almost all isolates sampled from potato cultures 3 belonged to AG-1, 7 to AG-3, and 2 to AG-7. RP1, RP2, RP3, and RP4 are special groups of R. solani in Saudi Arabia potato producing areas. Based on anastomosis group typing, cultural characterization and UP-PCR, we cannot arise clear conclusions about non-identified groups from potato. Further investigation of the population structure of R. solani collected from potato in Saudi Arabia is required to determine the extent and origin of the observed diversity. Utilization of molecular tool in combination with morphological characterization particularly when several samples were needed to distinguish.
Acknowledgement
This study was funded by the Department of Biology, Science and Humanities College, Alquwayiyah, Shaqra University, Saudi Arabia
References
- Abd-Elsalam K.A, Khalil M.S, Aly A.A, Asarn-Amal A. Genetic diversity among Fusarium oxysporum f. sp. vasinfectum isolates revealed by UP-PCR and AFLP markers. Phytopathol. Med. 2002;41:252-258.
- Abd-Elsalam K.A, Moslem M. A, Bahkali A.H. First Morpho-Molecular Identification of Rhizoctonia solani AG-7 from Potato tuber-borne sclerotium in Saudi Arabia. Afri. J. Microbiol. Res. 2009;3:952-956.
- Abd-Elsalam K.A, Guo J.R, Moslem M.A, Bahkali A.H, Verreet J.A. Suitability of intergenic spacer or internal transcribed spacer microsatellite-primed PCR for the identification of Rhizoctonia solani and some phytofungi. J. Rapid Aut. Methods Microbiol. 2009;17:383-397.
- Amaradasa B.S, Lakshman D, Horvath B.J, Amundsen K.L. Development of SCAR markers and UP-PCR cross-hybridization method for specific detection of four major subgroups of Rhizoctonia from infected turfgrasses. Mycologia. 2014;106:(1)163-172.
CrossRef - Banerjee S, Dutta S, Mondal N, Bhattacharyya S. Characterization of molecular variability in Rhizoctonia solani isolates from different agro-ecological zones by random amplified polymorphic DNA (RAPD) markers. Afri J. Biotechnol. 2012;11:9543-9548.
CrossRef - Bartz F.E, Cubeta M.A, Toda T, Naito S, Ivors K. An in planta method for assessing the role of basidiospores in Rhizoctonia foliar disease of tomato. Plant Dis. 2010;94:515-520.
CrossRef - Blazier S. R, Conway K. E. Characterization of Rhizoctonia solani isolates associated with patch diseases on turfgrasses. Proc. Okla. Acad. Sci. 2004;84:41-45.
- Carling D.E, Baird R.E, Gitaitis R.D, Brained K.A, Kuninaga S. Characterization of AG-13 a newly reported anastomosis group of Rhizoctonia solani. Dis .Control Pest Manage. 2002;9:893-899.
CrossRef - Carling D.E, Baird R.E, Gitaitis R.D, Brainard K.A, Kuninaga S. Characterization of AG-13 a newly reported anastomosis group of Rhizoctonia solani. Phytopathology. 2002;92:893-899.
- Cubeta M.A, Vilgalys R. Rhizoctonia. In: Encyclopedia of Microbiology Edited by: Lederberg J. San Diego: Academic Press. 2000;4:109-116.
- El-Hussieni S. Virulence of potato geographic isolates of Rhizoctonia solani (AG-s). Saudi J. Biol. Sci. 2004;11: 153-161.
- FAOSTAT. Food and Agriculture Organization of the United Nations, Statistics Division. Online: 2015. http://faostat3.fao.org.
- Fiers M, Edel-Hermann V, Heraud C, Gautheron N, Chatot C, Le Hingrat Y, Bouchek-Mechiche K, Steinberg C. Genetic diversity of Rhizoctonia solani associated with potato tubers in France. Mycologia. 2011;103:1230-44.
CrossRef - Guleria S, Aggarwal R, Thind T.S, Sharma T.R. Morphological and pathological variability in rice isolates of Rhizoctonia solani and molecular analysis of their genetic variability. J. Phytopathol. 2007;155:654-661.
CrossRef - KACST. King Abdulaziz City for science and technology. Strategic Priorities for Agricultural Technology Book. 2012;47.
- Kulik M.M, Dery P.D. Use of DAPI for anastomosis group typing of strains of the fungus. Rhizoctonia solani. Biotechnic Histochemistry. 1995;70:95–98. doi: 10.3109/10520299509108324
CrossRef - Lûbeck M, Lûbeck P.S. Universally primed PCR (UP-PCR) and its applications in mycology. In: Deshmukh S.K, Rai M.K, eds. Biodiversity of fungi: their role in human life. Enfield, New Hampshire: Science Publishers. 2005;409–438.
- Lûbeck M, Poulsen H. 2001. UP-PCR cross-blot hybridization as a tool for identification of anastomosis groups in the Rhizoctonia solani complex. FEMS Microbiol Lett. 201:83–89. doi:10.1016/S0378-1097(01)00245-2.
CrossRef - MacNish G.C, Carling D.E, Brainard K.A. Relationship of microscopic and macroscopic vegetative reactions in Rhizoctonia solani and the occurrence of vegetatively compatible populations (VCPs) in AG8. Mycol. Res. 1997;101:61–68.
CrossRef - Mishra P.K, Gogoi R, Singh P.K, Borah J, Rai S.N. Genotypic variability in isolates of Rhizoctonia solani from rice, maize and greengram. Indian Phytopath. 2015;68(1):56-62.
- Ogoshi A. Introduction: The genus Rhizoctonia. In: Sneh B, Jabaji-Hare S, Neate S, Dijst G, eds. Rhizoctonia Species. Taxonomy, Molecular Biology, Ecology, Pathology and Disease Control. Dordrecht, The Netherlands: Kluwer Academic. 1996;1-9.
CrossRef - Pathania N, Kanwar S.S, Jhang T, Koundal K.R, Sharmaer T.R. Application of different molecular techniques for deciphering genetic diversity among yeast isolates of traditional fermented food products of Western Himalayas. World J. Microbiol. Biotechnol. 2010;26:1539-1547.
CrossRef - Smiley R.W, Dernoeden P.H, Clarke B.B. Compendium of turfgrass diseases. St Paul, Minnesota: American Phytopathological Society. 2005.
- Sneh B, Burpee L, Ogoshi A. Identification of Rhizoctonia species. The APS, St. Paul, Minesota. 1998.
- Trigiano R.N, Ament M.H, Habera L.F, Caetano-Anolles G. Molecular techniques used to study systematics, ecology and evolution of plant pathogens. In: Trigiano RN, Windham MT, Windham AS, eds. Plant pathology concepts and laboratory exercises. Boca Raton, Florida: CRC Press. 2004;217–218.
- Truter M, Wehner F.C. Anastomosis grouping of Rhizoctonia solani associated with black scurf and stem canker of potato in South Africa. Plant Dis. 2004;88(83):2-83.2.
- Tsror L. Biology, Epidemiology and Management of Rhizoctonia solani on Potato. Journal of Phytopathology. 2010;158:649-658.
CrossRef - Wilson P.S, Ahvenniemi P.M, Lehtonen M.J, Kukkonen M, Rita H, Valkonen J.P.T. Biological and chemical control and their combined use to control different stages of of the Rhizoctonia disease complex on potato through the growing season. Annals of Applied Biology. 2008;153:307-320.
CrossRef - Woodhall J.W, Lees A.K, Edwards S.G, Jenkinson P. Characterization of Rhizoctonia solani from potato in Great Britain. Plant Pathology.2007;56:286-95.
CrossRef - Yugander A, Ladhalakshmi D, Prakasham V, Satendra K, Prasad M.S, Krishnaveni D, Madhav M.S, Sundaram, R.M, Laha G.S. Pathogenic and Genetic Variation among the Isolates of Rhizoctonia solani (AG 1-IA), the Rice Sheath Blight Pathogen. J. Phytopathol. 2015;163:465-474.
CrossRef

This work is licensed under a Creative Commons Attribution 4.0 International License.






