Manuscript accepted on : 12 June 2017
Published online on: --
Plagiarism Check: Yes
Farzana Gul, Anwar Hussain, Gul Jan and Muhammad Hamayun
Department of Botany, Abdul Wali Khan University Mardan, Pakistan.
Corresponding Author E-mail: hamayun@awkum.edu.pk
DOI : http://dx.doi.org/10.13005/bbra/2492
ABSTRACT: Presence of tough chitinous cell wall in fungi is a challenge for cell lysis and subsequent DNA isolation which has several downstream applications. We have developed an efficient protocol for extracting genomic DNA from endophytic fungi, which was isolated from different parts of Solanum xanthocarpum L. The procedure used was based on sodium dodecyl sulfate/ phenol/chloroform extraction method to remove polysaccharides and protein. The isolated genomic DNA was used to successfully amplify ITS region. The amplified ITS region was sequenced and homology of its sequence was used for confirming identity of the fungal endophyte. This rapid and inexpensive method can efficiently isolate high quality and quantity of genomic DNA suitable for PCR amplification and sequencing.
KEYWORDS: Genomic DNA extraction; endophytic fungi
Download this article as:| Copy the following to cite this article: Gul F, Hussain A, Jan G, Hamayun M. Genomic DNA Extraction for Molecular Identification of Endophytic Fungi: an Easy and Efficient Protocol. Biosci Biotech Res Asia 2017;14(2). |
| Copy the following to cite this URL: Gul F, Hussain A, Jan G, Hamayun M. Genomic DNA Extraction for Molecular Identification of Endophytic Fungi: an Easy and Efficient Protocol. Biosci Biotech Res Asia 2017;14(2). Available from: https://www.biotech-asia.org/?p=25829 |
Introduction
It has been anticipated that more than eighty percent of land plant species form symbiotic association with fungi.1 Fungal endophytes are those species that live inside tissues of living plant without harming their host.2 This symbiotic coexistence could have helped plant species adapt to new environmental stresses such as more extreme temperatures difference, increased exposure to solar radiation and desiccation.3 This association also stimulates productivity and plant growth in several crops.4 Thus fungal endophytes molecular characterization is an important step in this plant microbes interaction.5 DNA isolation is essential for the study of genome expression in fungus, because it is a requirement for many molecular biological techniques like construction of DNA libraries, gene isolation with polymerase quality and southern blot.6 Different protocols are used now a day for the isolation of genomic DNA from fungi.7 Though the quality and quantity of DNA, obtained through these protocols are generally acceptable, but these techniques are time consuming and costly.8 Therefore, the objective of the present study was to develop a rapid, easy and inexpensive method for the isolation of best quality of DNA from endophytic fungi.
Material and Methods
Endophytic Fungal Isolation
Solanum xanthocarpum was selected for the isolation of endophytic fungi. The whole plant parts were washed with tap water to remove dust particles, surface sterilized with 70% ethanol for 30 seconds and washed with double distilled water (ddH2O) to remove the surface contaminants. All parts were cut into 0.5 cm segments carefully placed in petri-plates containing Hagem media. The sterilized parts were also placed on separate Hagem plates to confirm the efficiency of surface sterilization. Emerging fungal hyphae were carefully picked and inoculated on fresh PDA media plates. The plates were incubated for 5 days at 28°C to obtain pure cultures of fully grown fungal colonies.
For DNA isolation, broth cultures of five fungal isolates were prepared. These isolates were selected on the basis of their high frequency of occurrence as endophytes in our plant. Flasks (250 mL) containing 50 mL Czapek-broth (Peptone 1%, Glucose, 1 %, 0.05% MgSO4.7H2SO4, 0.05 KCl and 0.001% FeSO4.7H2SO4: pH 7.3±0.2) were autoclaved and inoculated with fungal endophytes from PDA plates. The cultures in the flasks were then incubated on shaking incubator for 7 days at 28°C with a 120rpm. All the cultures were filtered separately to obtain the fungal biomass. The fresh fungal mycelium was used for fungal genomic DNA isolation.9
DNA Extraction Buffer and Solutions
Buffer used for DNA extraction from endophytes contained 0.5 mM hydroxymethyl-hydrochloride (Tris-HCl, pH 8.0), 0.5 mM ethylenediaminetetracetic acid, 0.1 mM NaCl and 5% SDS (Sodium dodisyl sulphate). Phenol/chloroform mixture was prepared by mixing equal volume of phenol and chloroform and the cloudy mixture was left overnight to get clear.10
Modified Protocol for DNA Extraction
For DNA isolation, previously reported method7 was followed. In short, 0.2g of fresh fungal sample was added in 500ul lysis buffer containing glass beads of different diameters. The sample was then vortex for about 10 min at maximum speed. No incubation was used in this method. The DNA obtained was checked for quality by running agarose gel electrophoresis.
In modified method, 200-500 mg of lyophilized fungal mycelium was crushed with the help of mortar and pestle in 500 μL of extraction buffer solution, containing 5% sodium dodecyl sulfate, 0.1M NaCl and 0.5M Tris-HCl, (pH8). After crushing, the grounded samples were shifted to 1.5 mL micro centrifuge tubes. The tubes were then vortex for 10-15 sec and then heated at 65°C for 30 min. during heating, the tubes were carefully inverted after every 5 min interval. After incubation, the tubes were centrifuged for 10 min at 11,000 g. supernatant were shifted to clean tubes and equal volume of chloroform, phenol, isoamyl alcohol mixture (24:25:1) was added. The tubes were vortex for 10-15 sec and then centrifuged for 5 min. at 10,000 g. The aqueous layer was taken in a new eppendorf tube. Chloroform: isoamyl alcohol (24:1) mixture was added in equal volume. The mixture was then centrifuged at 10,000 g for 5 min and supernatant obtained. The resultant supernatant was mixed with equal volume of absolute ethanol in a new Eppendorf to get the DNA precipitated. To improve the efficiency of precipitation, the Eppendorf containing mixture were incubated at 4ᵒC for 1h and then the precipitated DNA was separated from the mixture by centrifugation at 14,000 g for 10 min. Cold ethanol (70%) was used to wash the DNA pellets and the air dried pellets were taken in 40µL TE buffer. DNA solution was loaded onto agarose gel for electrophoresis and the gel was stained with 0.6 gm ethidium bromide to visualize DNA in the UV.
Quantity and Quality Determination
For quality assessment, the DNA samples were subjected to optical density (OD) analysis at 260 nm using Thermo Scientific Nano Drop 1000 spectrophotometer. Also, band quality on the agarose gel after electrophoresis was used as marker for DNA quality. Fitness of the isolated DNA for downstream application was checked by amplification of the ITS region.
PCR reaction was performed in a 20μL reaction volume following standard protocol.12 It consisted of 10 μl PCR master mix, 1μL of each ITS region universal primers (1/4) and 1μL DNA while the remaining volume was adjusted by adding 7ul ddH2O. The resultant product was gene cleaned using nucleogen gene clean kit and sequenced. The BLAST search program http://www.ncbi.nlm.nih.gov/BLAST/ was used to look for nucleotide sequence similar of 18S ITS (1/4) region for our fungal isolate. The obtained sequence was aligned by using ClustalW MEGA version 7.0 software for identification of our fungal endophyte.
Results and Discussion
Isolation and Characterization of the Endophytic Fungi
Solanum xanthocarpum plant was hosting 22 endophytic fungal isolates. Colonization frequency, isolation frequency, dominant fungi, endophytic infection rate of different fungal isolates are given in (Table 1). Isolation frequency of Solanum xanthocarpum is 72.9 % and endophytic infection rate is 67.57 %. The richest source of fungal endophytes were leaves and spines of S. xanthocarpum from which 7 and 8 isolates were isolated, respectively (Table 1). However, fungal diversity of the stem was low. Only 3 species were isolated. Spines were colonized by 8 fungal isolates each of which was a unique species making it a habitat of great fungal diversity among different parts studied. Species richness and colonization frequency is high in leaves and spines as compared to other parts of the plant. The richness of endophytes is higher in leaves and increases during leaf development, probably due to the changes in the structural, and chemical properties that accompany the leaf cycle.13 Phenolic content decreases as leaves mature, supporting diversity of endophytes. The colonization of endophytes can also change the leaf structure and pigment content and affect leaf physiology.14 Therefore several factors, associated with developmental stage, have a direct relationship with the diversity of endophytes in older leaves. Some scientists reported that these fungi generally exist in parenchyma, as well as the xylem and phloem. From xylem and phloem will move towards spines.15 That’s why spines have great fungal diversity.
Table 1: Fungal endophytes isolated from different parts of S. xanthocarpum along with their colonization frequency, dominance and richness
| Fungal strain | Root | Stem | Leaf | Spines | Fruit | Dominant fungi (%age) |
| SXL-A | – | 2 | 1 | – | – | 12 |
| SXL-B | – | – | 2 | – | – | 8 |
| SXL-C | – | – | 1 | – | – | 4 |
| SXL-D | – | – | 3 | – | 1 | 16 |
| SXL-E | – | – | 1 | – | – | 4 |
| SXL-F | – | – | 1 | – | – | 4 |
| SXR-G | 3 | 1 | – | 1 | – | 20 |
| SXR-H | 2 | – | – | – | – | 8 |
| SXR-I | 2 | – | – | – | – | 8 |
| SXR-J | 2 | – | 1 | – | – | 12 |
| SXR-K | 1 | – | – | – | – | 4 |
| SXSp-L | 1 | – | – | – | – | 4 |
| SXSp-M | – | – | – | 1 | – | 4 |
| SXSp-N | – | – | – | 1 | – | 4 |
| SXSp-O | – | – | – | 1 | – | 4 |
| SXSp-P | – | – | – | 1 | – | 4 |
| SXSp-Q | – | – | – | 1 | – | 4 |
| SXSp-R | – | – | – | 1 | – | 4 |
| SXSp-S | – | – | – | 2 | 1 | 12 |
| SXS-T | – | 1 | – | – | – | 4 |
| SXF-U | – | – | – | – | 1 | 4 |
| SXF-V | – | – | – | – | 1 | 4 |
| Total fungal isolates | 11 | 4 | 10 | 8 | 4 | – |
| Species richness | 6 | 3 | 7 | 8 | 4 | – |
| Colonization frequency | 86% | 43% | 88% | 100% | 57% | – |
DNA Isolation and Gel Electrophoresis
No DNA strands were visible after isolation steps, as indicated by established protocol.7 No clear bands were observed when DNA sample from reported method7 was run on gel electrophoresis, although glow appeared inside the wells.
The modified method revealed better result for DNA isolation, as clear from strands of DNA at the end of isolation protocol, as well as from gel electrophoresis. Yield of the DNA was high enough (60- 100 ng µL-1) to be suitable for downstream applications (Table 2). Genomic DNA isolated was of good quality yielding a single sharp band on 1% agarose gel (Figure 1).
Table 2: Quantity of genomic DNA isolated from dominant fungal endophytes isolated from S. xanthocarpum L.
| Fungal strain | DNA quantity in ng µL-1 |
| SXL-A | 60 |
| SXL-D | 70 |
| SXR-G | 100 |
| SXR-L | 60 |
| SXSp-S | 70 |
 |
Figure 1: DNA bands of fungal isolates SXL-A (A), SXL-D (D), SXR-G (G), SXR-L (L), SXSp-S (S). SXL (Solanum xanthocarpum leaf), SXR (Solanum xanthocarpum root), SXSp (Solanum xanthocarpum spines).
|
As the currently available fungal DNA extraction methods are relatively time consuming and costly,16 we developed a rapid DNA isolation protocol particularly suitable for low budget projects. Using this protocol, genomic DNA was isolated from five dominant fungal strains. The newly established protocol combines chemical and mechanical methods for digesting fungal chitenous cell wall to release genomic DNA of endophytes. Our method eliminates much of the time-consuming and laborious steps of other protocols.17 Reduction in vortex time and incubation in lysis buffer at 65C resulted in better isolation of DNA. High temperature treatment digest proteins, suspends lipids, and also digest/breakdown celluloid material making DNA.15
Sequencing of ITS Region and Identification of Endophytic Fungi
Sequences obtained from forward and reverse primers were aligned and the aligned sequence was subject to homology search by NCBI BLAST. Homology of the ITS region was used to identify the isolated endophyte as specie of Corynesporaa. Our sequence showed 99% homology with sequences of the ITS regions of Corynespora available in the Gene Bank database using the BLAST search program http://www.ncbi.nlm.nih.gov/BLAST.21
To check the suitability of the isolated genomic DNA in downstream molecular biology applications fungal strains were subjected to amplification by using universal primers for ITS region of 18 S rDNA18 (Van Burik et al., 1998). PCR is a basic protocol having a number of applications in molecular biology lab.19 Hence it is imperative for the isolated genomic DNA to be easily amplifiable by PCR which not only depend on the quality, but also on the quantity of the isolated DNA.20 Fungal genomic DNA isolated during current study was amicable to PCR amplification showing the absence of any inhibitor and also indicating its good quantity (Figure 1&2).
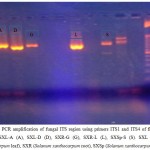 |
Figure 2: PCR amplification of fungal ITS region using primers ITS1 and ITS4 of five fungal strains; SXL-A (A), SXL-D (D), SXR-G (G), SXR-L (L), SXSp-S (S). SXL (Solanum xanthocarpum leaf), SXR (Solanum xanthocarpum root), SXSp (Solanum xanthocarpum spines).
|
References
- Al‐Samarrai T & Schmid J. A simple method for extraction of fungal genomic DNA. Letters in applied microbiology. 2000;30:53-56.
CrossRef - Bolano A., Stinchi S., Preziosi R., Bistoni F., Allegrucci M., Baldelli F., Martini A & Cardinali G. Rapid methods to extract DNA and RNA from Cryptococcus neoformans. FEMS yeast research. 2001;1:221-224.
- Drabkova L., Kirschner J & Vlĉek Ĉ. Comparison of seven DNA extraction and amplification protocols in historical herbarium specimens of Juncaceae. Plant Molecular Biology Reporter. 2002;20:161-175.
CrossRef - Gao T., Yao H., Song J., Liu C., Zhu Y., Ma X., Pang X., Xu H & Chen S. Identification of medicinal plants in the family Fabaceae using a potential DNA barcode ITS2. Journal of ethnopharmacology. 2010;130:116-121.
CrossRef - Hamayun M., Hussain A., Khan S. A., Irshad M., Khan A. L., Waqas M., Shahzad R., Iqbal A., Ullah N & Rehman G. Kinetin modulates physio-hormonal attributes and isoflavone contents of Soybean grown under salinity stress. Frontiers in plant science. 2015
- Kress W. J & Erickson D. L. A two-locus global DNA barcode for land plants: the coding rbcL gene complements the non-coding trnH-psbA spacer region. PLoS one. 2007;2.
- Kubicek C., Mach R., Peterbauer C & Lorito M. Trichoderma: from genes to biocontrol. Journal of Plant Pathology. 2001;11-23.
- Levin R. A., Wagner W. L., Hoch P. C., Nepokroeff M., Pires J. C., Zimmer E. A & Sytsma K. J. Family-level relationships of Onagraceae based on chloroplast rbcL and ndhF data. American Journal of Botany. 2003;90:107-115.
CrossRef - Li J., Zhao G. Z., Chen H. H., Wang H. B., Qin S., Zhu W. Y., Xu L. H., Jiang C. L & Li W. J. Antitumour and antimicrobial activities of endophytic streptomycetes from pharmaceutical plants in rainforest. Letters in applied microbiology. 2008;47:574-580.
CrossRef - Liu K., Ding X., Deng B & Chen W. Isolation and characterization of endophytic taxol-producing fungi from Taxus chinensis. Journal of industrial microbiology & biotechnology. 2009;36.
- Malinowski D. P & Belesky D. P. Adaptations of endophyte-infected cool-season grasses to environmental stresses: mechanisms of drought and mineral stress tolerance. Crop Science. 2000;40:923-940.
CrossRef - Peer W. A & Murphy A. S. Flavonoids and auxin transport: modulators or regulators? Trends in plant science. 2007;12:556-563.
- Płaza G., Upchurch R., Brigmon R., Whitman W & Ulfig K. Rapid DNA extraction for screening soil filamentous fungi using PCR amplification. Polish Journal of Environmental Studies. 2004;13:315-318. doi: 10.15244. Kraj, PL.
- SAMBROOK J & RUSSELL D. W. Molecular cloning: a laboratory manual 3rd edition. Coldspring-Harbour Laboratory Press, UK. 2001.
- SMITH S. E & READ D. J. Mycorrhizal symbiosis, Academic press. 2010.
- VAN BURIK J. A., MYERSON D., SCHRECKHISE R. W & BOWDEN R. A. Panfungal PCR assay for detection of fungal infection in human blood specimens. Journal of Clinical Microbiology. 1998;36:1169-1175.
- VOLLMER F., ARNOLD S., BRAUN D., TERAOKA I & LIBCHABER A. Multiplexed DNA quantification by spectroscopic shift of two microsphere cavities. Biophysical journal. 2003;85:1974-1979.
CrossRef - Waqas M., Khan A. L., Kamran M., Hamayun M., Kang S. M., Kim Y. H & Lee I. J. Endophytic fungi produce gibberellins and indoleacetic acid and promotes host-plant growth during stress. Molecules. 2012;17:10754-10773.
CrossRef - ZHANG J & STEWART J. Economical and rapid method for extracting cotton genomic DNA. J Cotton Sci. 2000;4:193-201.

This work is licensed under a Creative Commons Attribution 4.0 International License.





