Manuscript accepted on : 19 October 2016
Published online on: --
Plagiarism Check: Yes
Molecular Characterization of Lactic Acid Bacteria Isolated from Rumen Liquor of Goat
Gothandam Ladha and Kadirvelu Jeevaratnam*
Department of Biochemistry and Molecular Biology, Pondicherry University, R.V.Nagar, Kalapet-605 014, India.
Corresponding Author E-mail: jeevskj@gmail.com
DOI : http://dx.doi.org/10.13005/bbra/2378
ABSTRACT: Objective of this study is to isolate and molecular characterise lactic acid bacteria from rumen liquor of goat. A total of 58 isolates, 42 bacilli and 16 cocci on gram staining were obtained. Screening for antimicrobial activity using cell free supernatant resulted in 12 potentisolates, 9 bacilli and 3 cocci exhibiting wide range spectrum of antimicrobial activity against food borne pathogens. The antimicrobial activity was lost on treatment with protease suggesting the isolates bacteriocinogenic in nature. These potent bacteriocinogenic isolates were characterized by physiological and biochemical properties including carbohydrate utilization profile and RAPD profile using M13 and R2 primers to differentiate at strain level. The 16 rRNA gene sequence analysis coupled with multiplex PCR analysis using recA gene amplification ther isolates were identified as Lactobacillus plantarum (7), Lactobacillus fermentum (2) and Pediococcus pentosaceus (3). Further work on their probiotic and other beneficial properties may lead to several applications and benefit to animal and humans.
KEYWORDS: Rumen liquor; Pediococcus; Lactobacillus; RAPD; 16S rRNA
Download this article as:| Copy the following to cite this article: Ladha G, Jeevaratnam K. Molecular Characterization of Lactic Acid Bacteria Isolated from Rumen Liquor of Goat. Biosci Biotech Res Asia 2016;13(4). |
| Copy the following to cite this URL: Ladha G, Jeevaratnam K. Molecular Characterization of Lactic Acid Bacteria Isolated from Rumen Liquor of Goat. Biosci Biotech Res Asia 2016;13(4). Available from: https://www.biotech-asia.org/?p=17077 |
Introduction
Lactic acid bacteria (LAB) are a group of Gram-positive, non-spore forming, non-respiring bacilli or cocci, catalase negative, anaerobic, acid tolerant and strictly fermentative producing lactic acid. LAB are widely distributed in nature. LAB can be isolated from grains, dairy and meat products, fermenting vegetables and mucosal surface of animals1. The LAB show special promise for selection and use as protective cultures in various food systems. They are also considered for biological preservation of foods2. The preservative factors produced by LAB have been identified as antimicrobial small proteins/peptides apart from lactic acid, acetic acid, etc. LAB can also be employed as probiotics to protect from food or feed borne infectious organisms. LAB play significant role in the maintenance of gastrointestinal tract of both human and animals and improve digestive function3,4.
Rumen liquor is a concentrated liquid found in the rumen of an animal consisting of chewed up feed with large amount of saliva and microorganisms, mainly bacteria and protozoa. It is used to test the digestability of feed or the nutrient balance of an animal’s diet. In the digestive tract of cattle, lactic acid bacteria (LAB) were found in the rumen5. Thus, the objective of this study is to isolate and identify LAB from rumen liquor of commercially and widely consumed goats and screen for the antimicrobial potential towards food borne pathogens.z
Materials and Methods
Isolation of LABs from rumen liquor
The rumen liquor sample was collected from slaughter house as soon as animal wassacrificed, kept in sterile saline solution and then transferred to laboratory. The LAB were isolated by serial dilution and spread plate technique in LAB selective media, de Man Rogosa Sharpe agar (MRSA) at 37°C for 48 h in anaerobic conditions. The colonies on MRS agar which were milky white, circular, convex, elevated and non-pigmented were chosen. The pure cultures were obtained by quadrant streaking on MRS agar and preserved in -20°C using glycerol as cryoprotectant.
Screening of isolates for antimicrobial activity against food pathogens
The cell free supernatant concentrate (CFSC; 10-fold) of LAB isolates was adjusted to pH 5.0 using NaOH (3 M) and evaluated for antimicrobial activities against various food pathogens such as Staphylococcus aureus MTCC 3160, Bacillus cereus MTCC-1272, Listeria monocytogenes MTCC-1143, Escherichia coli MTCC-1687, Salmonella typhi MTCC-734 and Serratia marcescens MTCC-97 by agar-well diffusion method6. The zone of inhibition was measured after 24 h. In order to determine, whether the antibacterial activity was due to a proteinaceous substance, the protease treated CFSC was also used for antibacterial activity against L. monocytogenes7.
Physiological, Biochemical and Molecular characterisation of potent isolates
Physiological and biochemical characterization of isolates
The potent isolates were primarily characterized based on Bergey’s manual of systematic bacteriology8. The potent bacterial isolates were tested for Gram’s characteristics, catalase, and growth at different temperature (15, 37 and 45°C), NaCl concentrations (4, 6.5, and 10%), gelatin hydrolysis, starch hydrolysis, acetion production, ammonia production, gas formation, slime production, homo-hetero fermentation using standard protocols and carbohydrate utilization profile using Hi-Carbohydrate kit (Hi-Media, India).
RAPD analysis
The genomic DNA of potent bacterial isolates was isolated from overnight cultures of potent isolates9 and the purity was checked spectrophotometrically (Shimadzu, Japan). Genomic DNA was used for RAPD analysis, using M13 5’-GAGGGTGGCGGTTCT-3’ and R2 5’-GGCGACCACTAG-3’ primers as described10,11. PCR was performed with 50µl reaction mixture containing 1X buffer, 1.5mmol l-1MgCl2, 200µmol l-1 of dNTPs mix, 0.5µmol l of primer (Eurofins Genomics, India Pvt. Ltd., India), 50ng of genomic DNA , and 1U of Taq DNA polymerase (Merck Biosciences, India). The DNA was amplified with Mastercycler gradient (Eppendorf, Germany) with initial denaturation at 94°C for 5 min, 30 cycles of 94°C for 40 s, annealing at 40°C (M13) and 38°C for (R2) followed by ramping to 72°C for 0.5°C/s and extension for 2 min with final extension at 72°C for 5min. The products were checked using 1.5% agarose gel electrophoresis with Tris Borate EDTA (TBE) buffer at 100 V and stained with ethidium bromide.
16S rRNA gene sequence and phylogenetic analysis
The 16S r RNA gene was amplified for the selected isolates using universal primer UIF 5’AGAGTTTGATCCTGGCTCAG3’ and U1R 5’GGTTACCTTGTTACGACTT 3’ as described12. The PCR cocktail (50 μL) consisting of primers (50 pM) , (Sigma Aldrich, India)genomic DNA (50 ng), Taq DNA buffer (1X), MgCl2 (3 mM), dNTP (0.4 mM), and Taq polymerase (1U/μL) was used for amplification under the conditions of initial denaturation at 94°C for 5min, 30 cycles of denaturation at 94°C for 1 min, annealing at 55°C for 1min and extension at 72°C for2 min and a final extension at 72°C for 10 min. The product was run on agarose gel electrophoresis (1%) with TBE buffer and stained with ethidium bromide. The 1.5 kb PCR product was cleaned using clean-up kit (Merck Bioscience, India) and sequenced at Eurofines Genomics India, Pvt Ltd., Bangalore, India. BLASTn was done and reference sequences were collected from NCBI and aligned using ClustalW. The phylogenetic tree was constructed by neighbour-joining (NJ) method with MEGA 5.0 software after resampling 1000 times with bootstrap analysis. Bacillus cereus was used as an out group. Multiplex PCR analysis was also carried out on bacilli isolates13.
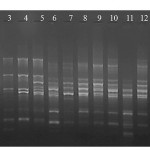 |
Figure 1: M13 primer RAPD fingerprinting profile of selected 12 isolates from goat rumen. Where Lane M – Marker, Lane 1 – LJR2, Lane 2 – LJR 3, Lane 3 – LJR 1, Lane 4 – LJR 5, Lane 5 – LJR 9, Lane 6 – LJR 7, Lane 7 – LJR 8, Lane 8 – LJR 13, Lane 9 – LJR18, Lane 10 – LJR 4, Lane 11 – LJR6 and Lane 12 – LJR10. |
Nucleotide accession numbers
The nucleotide sequences of the isolates were deposited in GenBank having accession numbers KT864675 to KT864683; KX085413 to KX085415.
Result and Discussion
Present study deals with the isolation and characterization of LAB from rumen liquor of goat. A total of 58 isolates were isolated from rumen of goat based on their ability to on MRS agar. Gram staining of all the isolates showed 42 gram positive bacilli and 16 gram positive cocci. Screening of CFSC of the isolates for antimicrobial activity against food borne pathogens showed 12 isolates (9 bacilli, viz., LJR2, LJR3, LJR4, LJR6, LJR7, LJR8, LJR10, LJR13 and LJR18 and 3 cocci, viz., LJR1, LJR5 and LJR9 to have potent antibacterial activity (Table 1). Acid protease treatment of the CFSC showed loss of activity inferring bacteriocinogenic nature of the isolates. All the isolates are able to grow at temperature 15 to 45 oC, pH at 4.5 to 7.5 while no growth at pH 3.5; and at salt concentration of 4 and 6.5% with no growth at 10% NaCl. All the isolates are homo-fermentative indicating production of lactic acid as major end product; lactic acid produced found to be of DL configuration. All isolates showed positive for esculin hydrolysis, while negative for starch, gelatine, arginine and casein hydrolysis; negative for production of catalase and acetoin and slime from sucrose. Carbohydrate utilization profile for all the isolates is provided in Table 4. All isolates utilize dextrose and fructose, while negative for citrate, malonate, sorbose, ONPG, rhamnose, alpha methyl-D-glucoside and manitol. The isolates showed a differential response for all other sugars (Table 2). Physiological and biochemical characteristics depicted the isolates LJR5 and LJR9 are very much similar, however, RAPD analysis using M13 and R2primers (Fig. 1 & 2) led to 10 groupings and also differentiated these two isolates, LJR5 and LJR9. Finally, all the 12 isolates are found to be different at strain level.
 |
Table 1: Antibacterial activity of LAB isolates against food pathogens |
 |
Table 2: Sugar utilization profile of the LAB isolates. |
The 16S rRNA gene sequence analysis of all the 7 bacilli isolates, viz., LJR2, LJR3, LJR4, LJR7, LJR8, LJR10 and LJR13 revealed 99% homology with L. pentosus, L. plantarum and L. paraplantarum using BLASTn of NCBI and the phylogenetic tree constructed based on Neighbour – joining method (Fig. 2), and other 2 bacilli, viz., LJR6 and LJR18 belong to L. fermentum, while the 3 cocci, viz., LJR1, LJR5 and LJR9 belong to Pediococcus pentosaceus. As L. pentosus, L. plantarum and L. paraplantarum are genotypically closely related (>97%), it was difficult to differentiate them using16S ribosomal DNA sequence14,15; the 7 Lactobacillus strains were further confirmed at the species level by multiplex PCR based on recA gene sequence. Multiplex PCR assay gave the amplification product of 318 bp for all the 7 isolates confirming that they belong to L. plantarum13.
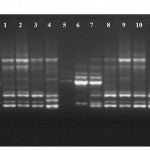 |
Figure 2: R2 primer RAPD fingerprinting profile of selected isolates. Lane M – Marker, Lane 1 – LJR2, Lane 2 – LJR3, Lane 3 – LJR7, Lane 4 – LJR8, Lane 5 – LJR1, Lane 6 – LJR5, Lane 7 – LJR9, Lane 8 – LJR13, Lane 9 – LJR18, Lane 10 – LJR4, Lane 11 – LJR6 and Lane 12 – LJR10.
|
 |
Figure 3: Phylogenetic tree constructed with neighbour joining method using Mega 5.0 software showing the position of LJR isolates from rumen liquor.
|
In conclusion, the Rumen liquor of goat harbours LAB of both Lactobacillus and Pediococcus genus exhibiting wide spectrum of activity against food pathogens due to production of bacteriocins. Thus, these potent bacteriocinogenic isolates have to be evaluated further for its probiotic and other beneficial properties
Acknowledgements
The authors acknowledge UGC – SAP and DST – FIST programme sponsored instrument facilities. Author, Ms. G. Ladha is also thankful to University Grants Commission, New Delhi for financial assistance No.F.7-370/ 2012 through BSR Fellowship.
References
- Lindgren, S. W., Dobrogosz, W., J. Antagonistic activities of lactic acid bacteria in food and feed fermentations. FEMS Microbial Rev. 87:149-164 (1990).
CrossRef - Ganguly, S. Biologically viable methods for food preservation: A review. Res J Chem Environ. 1: 01-02 (2013).
- Kim, P.I., Jung, M.Y., Chan, Y.H., Kim, S., Kim, S.J., Park, Y.H. Probiotic properties of lactobacillus and Bifidobacterium strains isolated from Porcine Gastrointestinal tract. Appl Microbiol Biotechnol. 74: 1103-1111 (2007).
CrossRef - Marco, M.L., Pavan, S., Kleerebezem, M. Towards understanding molecular modes of probiotic action. Curr Opin Biotechnol. 17: 204-210 (2006).
CrossRef - Cobos, M. A., de Coss, A. L., Ramirez, N. D., Gonzalez, S. S.,Cerrato, R. F. Pediococcus acidilactici isolated from the rumen of lambs with rumen acidosis, 16S rRNA identification and sensibility to monensin and lasalocid. Res Vet Sci. 90: 26-30 (2011).
CrossRef - Tagg, J. R., McGiven, A. R. Assay systems for bacteriocins. Appl Microbiol. 21: 125 (1971).
- Moraes, P.M., Perin, L.M., Ortolani, M.B.T., Yamazi, A. K., Vicosa, G.N., Nero, L. A. Protocols for the isolation and detection of lactic acid bacteria with bacteriocinogenic potential. LWT-Food Sci Technol. 43: 1320-1324 (2010).
CrossRef - Krieg, N. Bergey’s manual of systematic bacteriology, Vol 2. Williams and Wilkins, Baltimore: (1984).
- de Los Reyes-Gavilan, C.G., Limsowtin, G.K., Tailliez, P., Sechaud, L., Accolas, J.P. A Lactobacillus helveticus-specific DNA probe detects restriction fragment length polymorphisms in this species. Appl Environ Microbiol. 58:3429-3432 (1992).
- Rossetti, L., Giraffa, G. Rapid identification of dairy lactic acid bacteria by M13 – generated, RAPD-PCR fingerprint databases. J Microbiol Meth. 63: 135-144 (2005).
CrossRef - Bonomo, G. M., Ricciardi, A., Zotta, T., Parente, E., Salzano, G. Molecular and technological characterization of lactic acid bacteria from traditional fermented sausages of Basilicata region (Southern Italy). Meat Sci. 80: 1238-1248 (2008).
CrossRef - Singh, A. K., Ramesh A. Evaluation of a facile method of template DNA preparation for PCR – based detection and typing of lactic acid bacteria. Food Microbiol.26: 504-513. (2009).
CrossRef - Torriani, S., Felis, G. E., Dellaglio, F. Differentiation of Lactobacillus plantarum, pentosus, and L. paraplantarum by recA gene sequence analysis and multiplex PCR assay with recA gene – derived primers. Appl Environ Microbiol. 67: 3450-3454 (2001).
CrossRef - Felis, G., Dellaglio, F. Taxonomy of lactobacilli and bifidobacteria. Curr Issues Intest Microbiol. 8: 44-61 (2007).
- Singh, S., Goswami, P., Singh, R., Heller, K. J. Application of molecular identification tools for Lactobacillus, with a focus on discrimination between closely related species: A review. LWT–Food Sci Technol. 42: 448-457 (2009).
CrossRef

This work is licensed under a Creative Commons Attribution 4.0 International License.





