Manuscript accepted on : 15 August 2016
Published online on: --
Plagiarism Check: Yes
Electrophoretic Analysis of Seed Proteins of Rice Varieties
Innabat A. Sartbayeva1,2, Bakdaulet N. Usenbekov1, Aiman B. Rysbekova1, Kulpash M. Bulatova3, Dauren T. Kazkeyev1, Eldos A. Zhanbyrbaev1, Horlan A. Berkimbay1 and Kabyl Zh. Zhambakin1
1Institute of Plant Biology and Biotechnology (Almaty 050040, Kazakhstan).
2Al-Farabi Kazakh National University (Almaty 050040, Kazakhstan).
3LLP Kazakh Scientific and research Institute of Land Farming and Plant Growing, Almalybak.
Corresponding Author E-mail: bakdaulet7@yandex.ru
DOI : http://dx.doi.org/10.13005/bbra/2349
ABSTRACT: Composition of seed storage proteins were evaluated in rice collection materials using electrophoresis in alkaline medium at presence of SDS. Optimal conditions for protein extraction and fractionation were determined, the classification of protein components spectrum and methods for composition protein formulation was developed. It was conducted the registration and clustering of 62 rice samples, of which 5 were heterogeneous in gel electrophoresis of electropherograms.
KEYWORDS: rice; SDS-PAGE; storage protein; relative electrophoretic mobility
Download this article as:| Copy the following to cite this article: Sartbayeva I. A, Usenbekov B. N, Rysbekova A. B, Bulatova K. M, Kazkeyev D. T, Zhanbyrbaev E. A, Berkimbay H. A, Zh. Zhambakin K. Electrophoretic Analysis of Seed Proteins of Rice Varieties. Biosci Biotech Res Asia 2016;13(4). |
| Copy the following to cite this URL: Sartbayeva I. A, Usenbekov B. N, Rysbekova A. B, Bulatova K. M, Kazkeyev D. T, Zhanbyrbaev E. A, Berkimbay H. A, Zh. Zhambakin K. Electrophoretic Analysis of Seed Proteins of Rice Varieties. Biosci Biotech Res Asia 2016;13(4). Available from: https://www.biotech-asia.org/?p=16584 |
Introduction
Polymorphism of the reserve proteins of seeds is widely used in the identification of crops varieties, evaluation of the genetic diversity of collections, marking of agronomic characters [1, 2]. Content of protein in the corn and grits of rice is relatively low (6-15% and 5-11%, respectively), but its balanced composition with high content of the very important essential lysine amino acid causes high nutritional value and digestibility of the rice products [3, 4].
Most of the reserve proteins is accounted for by glutelin proteins – oryzenins (up to 80% of the total protein content in the grain), while salt- and alcohol-soluble proteins are represented in much smaller numbers [5]. Alcohol-soluble prolamins are less than 5% of total protein. Lack of alcohol-soluble proteins reduces the possibility of allergic reactions and disease of atrophy of the gastrointestinal tract and celiac disease at use of rice derivative products in food.
Investigation of composition of the rice reserve proteins by different groups of researchers has shown that the most heterogeneous protein spectrum was obtained through fractioning them in the alkaline medium in the presence of sodium dodecyl sulfate. Researchers have various opinions regarding the rice polymorphism on the composition of glutenin and prolamin. Thus, in a number of publications it indicates that the composition of proteins in different genotypes of rice is similar, with some exceptions, whereas other authors, on the contrary, reveal considerable variability in the protein spectrum [6, 7]. Clear difference is revealed in the profile of albumin and glutelins of lines O. sativa and collectible samples of wild forms of O.glumaepatula, the latter can be used in breeding for improving the nutritional value of reserve proteins [10]. Electrophoresis of reserve proteins is informative for breeding because certain components of protein spectrum, their proportions, and intensity of manifestations in the spectrum can serve as markers for rice indicators important in breeding: the genetic purity of varieties, differentiability of lines, low or high content of amylose in the grain, presence of a reducing gene [11, 12, 13, 14].
Objective
Adapting the methods of extraction and electrophoresis of storage proteins in rice varieties and collectible samples, and identifying the suitability of the method of protein markers in their classification.
Materials and Methods
The objects of research were released varieties of rice breeding of the Institute of plants biology and biotechnology, Kazakh Research Institute of Rice named after I. Zhahaev (Kyzylorda), as well as varieties and collectible samples of the world breeding in amount of 62 from the breeding of the All-Russian Rice Research Institute (Krasnodar, Russia).
Storage proteins were extracted by means of Tris-HCl and phosphate buffer pH 6.8, containing DDSNa, b-mercaptoethanol, glycerine and bromophenol blue dye. Fractionation was performed in polyacrylamide gel of 10 and 12% concentration by the modified Laemmli method [15]. Mathematical data processing was carried out by WARD clustering method by the program Statistica V 5.0.
Results and discussion
Comparison of extraction of storage proteins by different buffer systems and separation in polyacrylamide gels of various densities showed that the most clear distinction with the manifestation of all the groups of proteins (glutelin, prolamin, albumin and globulins) was observed at use of the extracting solution for the extraction of proteins on the basis of phosphate buffer and fractionation in 12% polyacrylamide gel.
The storage proteins spectrum of rice grains produced in our fractionation conditions consisted of 26-28 components with varying molecular weight from 136 to 15 kD (Figure 1). Registration of protein components was carried out by the relative electrophoretic mobility (REM) in the protein spectrum divided into: area A of slowly moving subunits with REM from 7 to 40, area B with components REM from 44 to 64, and area C of fast moving proteins with REM from 70 to 90. Table 1 shows the protein formula of the analyzed seeds of 62 rice varieties. The analysis results showed that components of area A are the most informative (differentiating components are indicated by arrows), while components of areas B and C are less variable, similar in the number, intensity and REM.
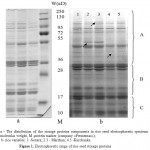 |
Figure 1: Electrophoretic range of rice seed storage proteins
|
Some varieties, such as Bakanasski, Priozerny 61, Lider, Madina and collectible sample 30-09 were heterogeneous in composition of storage proteins.
Table 1: The composition of storage proteins collectible varieties of rice
| Varieties and rice samples of collection | REM components, zone | ||||||||||||||||||||||||||||||||
|
А zone |
В zone | С zone | |||||||||||||||||||||||||||||||
| # | 1 | 2 | 3 | 4 | |||||||||||||||||||||||||||||
| REM | 6 | 7 | 8 | 10 | 11 | 14 | 15 | 19 | 24 | 26 | 28 | 30 | 31 | 35 | 40 | 44 | 45 | 47 | 52 | 55 | 60 | 64 | 70 | 72 | 73 | 77 | 80 | 84 | 87 | 90 | |||
| 1 | Viola | – | 2 | – | 1 | 2 | 1 | 1 | 1 | 3 | 1 | 1 | 1 | 1 | 3 | 3 | 4 | 2 | 1 | 1 | 2 | 1 | 3 | 3 | 1 | 1 | 2 | 3 | 3 | ||||
| 2 | Kuban 3 | 2 | – | 1 | 2 | 1 | 1 | 1 | 3 | 1 | 1 | 1 | 1 | 3 | 3 | 4 | 2 | 1 | 1 | 2 | 1 | 3 | 3 | 1 | 1 | 2 | 3 | 3 | |||||
| 3 | Bakanasski 1 | 2 | – | 2 | 1 | 2 | 1 | 1 | 1 | 3 | 3 | 2 | 1 | 1 | 1 | 1 | 3 | 3 | 4 | 2 | 1 | 1 | 2 | 1 | 3 | 3 | 1 | 1 | 2 | 3 | 3 | ||
| Bakanasski 2 | 2 | – | 2 | 1 | 2 | 1 | 1 | 1 | – | 3 | 2 | 1 | 1 | 1 | 1 | 3 | 3 | 4 | 2 | 1 | 1 | 2 | 1 | 3 | 3 | 1 | 1 | 2 | 3 | 3 | |||
| 4 | Sonata | – | 2 | 1 | 1 | 2 | 1 | 1 | 1 | 3 | 1 | 1 | 1 | 1 | 3 | 3 | 4 | 2 | 1 | 1 | 2 | 1 | 3 | 3 | 1 | 2 | 2 | 3 | 3 | ||||
| 5 | Marzhan | 2 | – | 2 | 1 | 2 | 1 | 1 | 1 | 3 | 3 | 2 | 1 | 1 | 1 | 1 | 3 | 3 | 4 | 2 | 1 | 1 | 2 | 1 | 3 | 3 | 1 | 1 | 2 | 3 | 3 | ||
| 6 | Kurchanka | – | 2 | 1 | 1 | 2 | 1 | 1 | 1 | 3 | 1 | 1 | – | 1 | 1 | 3 | 3 | 4 | 2 | 1 | 1 | 2 | 1 | 3 | 3 | 1 | 1 | 2 | 3 | 3 | |||
| 7 | Darii 23 | 2 | 1 | – | 1 | 2 | 1 | 1 | 1 | 3 | 2 | 1 | – | 1 | 1 | 3 | 3 | 4 | 2 | 1 | 1 | 2 | 1 | 3 | 3 | 1 | 2 | 2 | 3 | 3 | |||
| 8 | Analog 2 | – | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 3 | 1 | 1 | 1 | 1 | 3 | 3 | 4 | 2 | 1 | 1 | 2 | 1 | 3 | 3 | 1 | 1 | 2 | 3 | 3 | ||||
| 9 | Sample 34-09 | 1 | 1 | – | 1 | 2 | 1 | 1 | 1 | 3 | 1 | 1 | 1 | 1 | 3 | 3 | 4 | 2 | 1 | 1 | 2 | 1 | 3 | 3 | 1 | 1 | 2 | 3 | 3 | ||||
| 10 | Hankaisky 429 | – | 2 | 1 | 1 | 2 | 1 | 1 | 1 | 3 | 1 | 1 | 1 | 1 | 3 | 3 | 4 | 2 | 1 | 1 | 2 | 1 | 3 | 3 | 1 | 1 | 2 | 3 | 3 | ||||
| 11 | Liman | – | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 3 | 1 | 1 | 1 | 1 | 3 | 3 | 4 | 2 | 1 | 1 | 2 | 1 | 3 | 3 | 1 | 1 | 2 | 3 | 3 | ||||
| 12 | Sample 49-09 | 1 | 1 | – | 1 | 2 | 1 | 1 | 1 | 3 | 2 | 1 | – | 1 | 1 | 3 | 3 | 4 | 2 | 1 | 1 | 2 | 1 | 3 | 3 | 1 | 2 | 2 | 3 | 3 | |||
| 13 | Black rice | 2 | 1 | – | 1 | 2 | 1 | 1 | 1 | 3 | 2 | 1 | – | 1 | 1 | 3 | 3 | 4 | 2 | 1 | 1 | 2 | 1 | 3 | 3 | 1 | 2 | 2 | 3 | 3 | |||
| 14 | Lebed | – | 2 | 1 | 1 | 2 | 1 | 1 | 1 | 3 | 2 | 1 | 1 | 1 | 1 | 3 | 3 | 4 | 2 | 1 | 1 | 2 | 1 | 3 | 3 | 1 | 2 | 2 | 3 | 3 | |||
| 15 | Ametist | 2 | 2 | – | 1 | 2 | 1 | 1 | 1 | 2 | 2 | 1 | 1 | 1 | 3 | 3 | 4 | 2 | 1 | 1 | 2 | 1 | 3 | 3 | 1 | 2 | 2 | 3 | 3 | ||||
| 16 | ARRI 10173 | 1 | 1 | – | 1 | 2 | 1 | 1 | 1 | 2 | 2 | – | 1 | 1 | 1 | 3 | 3 | 4 | 2 | 1 | 1 | 2 | 1 | 3 | 3 | 1 | 1 | 2 | 3 | 3 | |||
| 17 | ARRI 10177 | – | 2 | 1 | 1 | 2 | 1 | 1 | 1 | 2 | 3 | 1 | 1 | 1 | 1 | 1 | 3 | 3 | 4 | 2 | 1 | 1 | 2 | 1 | 3 | 3 | 1 | 1 | 2 | 3 | 3 | ||
| 18 | ARRI 10178 | 2 | – | 2 | 1 | 2 | 1 | 1 | 1 | 3 | 3 | 1 | – | 1 | 1 | 1 | 3 | 3 | 4 | 2 | 1 | 1 | 2 | 1 | 3 | 3 | 1 | 1 | 2 | 3 | 3 | ||
| 19 | Fisht | 2 | 2 | – | 1 | 2 | 1 | 1 | 1 | 3 | 1 | – | 1 | 1 | 1 | 3 | 3 | 4 | 2 | 1 | 1 | 2 | 1 | 3 | 3 | 1 | 2 | 2 | 3 | 3 | |||
| 20 | Deti Vetra | 2 | 1 | – | 1 | 2 | 1 | 1 | 1 | 2 | 3 | 2 | 1 | 2 | 1 | 1 | 3 | 3 | 4 | 2 | 1 | 1 | 2 | 1 | 3 | 3 | 1 | 1 | 2 | 3 | 3 | ||
| 21 | Lugovoi | 1 | 1 | – | 1 | 2 | 1 | 1 | 1 | 3 | 1 | – | 1 | 1 | 1 | 3 | 3 | 4 | 2 | 1 | 1 | 2 | 1 | 3 | 3 | 1 | 1 | 2 | 3 | 3 | |||
| 1 | 2 | 3 | 4 | ||||||||||||||||||||||||||||
| 22 | Priozerny 61. 1 | 2 | 2 | – | 1 | 2 | 1 | 1 | 1 | 3 | 2 | 1 | 1 | 1 | 1 | 3 | 3 | 4 | 2 | 1 | 1 | 2 | 1 | 3 | 3 | 1 | 2 | 2 | 3 | 3 | |
| Priozerny 61. 2 | 2 | 2 | – | 1 | 2 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 3 | 3 | 4 | 2 | 1 | 1 | 2 | 1 | 3 | 3 | 1 | 2 | 2 | 3 | 3 | ||
| 23 | Sample 1-09 | 2 | 1 | – | 1 | 2 | 1 | 1 | 1 | 3 | 1 | – | 1 | 1 | 1 | 3 | 3 | 4 | 2 | 1 | 1 | 2 | 1 | 3 | 3 | 1 | 1 | 2 | 3 | 3 | |
| 24 | Sample 4-09 | 1 | – | 2 | 1 | 2 | 1 | 1 | 1 | 3 | 1 | – | 1 | 1 | 1 | 3 | 3 | 4 | 2 | 1 | 1 | 2 | 1 | 3 | 3 | 1 | 1 | 2 | 3 | 3 | |
| 25 | Sample 9-09 | 2 | – | 2 | 1 | 2 | 1 | 1 | 1 | 2 | 3 | 1 | – | 1 | 1 | 1 | 3 | 3 | 4 | 2 | 1 | 1 | 2 | 1 | 3 | 3 | 1 | 1 | 2 | 3 | 3 |
| 26 | Sample 10-09 | 1 | 1 | – | 1 | 2 | 1 | 1 | 1 | 3 | 1 | – | 1 | 1 | 1 | 3 | 3 | 4 | 2 | 1 | 1 | 2 | 1 | 3 | 3 | 1 | 1 | 2 | 3 | 3 | |
| 27 | Sample 29-09 | 1 | 1 | – | 1 | 2 | 1 | 1 | 1 | 3 | 2 | 1 | – | 1 | 1 | 1 | 3 | 3 | 4 | 2 | 1 | 1 | 2 | 1 | 3 | 3 | 1 | 2 | 2 | 3 | 3 |
| 28 | Sample 30-09 1 | 2 | 1 | 1 | 2 | 1 | 1 | 1 | 2 | 3 | 1 | 2 | 1 | 1 | 1 | 3 | 3 | 4 | 2 | 1 | 1 | 2 | 1 | 3 | 3 | 1 | 2 | 2 | 3 | 3 | |
| Sample 30-09 2 | 2 | 1 | 1 | 2 | 1 | 1 | 1 | 2 | 3 | 1 | 1 | 1 | 1 | 1 | 3 | 3 | 4 | 2 | 1 | 1 | 2 | 1 | 3 | 3 | 1 | 1 | 2 | 3 | 3 | ||
| 29 | Sample 32-09 | 1 | – | 2 | 1 | 2 | 1 | 1 | 1 | 3 | 1 | – | 1 | 1 | 1 | 3 | 3 | 4 | 2 | 1 | 1 | 2 | 1 | 3 | 3 | 1 | 2 | 2 | 3 | 3 | |
| 30 | Sample 46-09 | 1 | 1 | – | 1 | 2 | 1 | 1 | 1 | 3 | 2 | – | 1 | 1 | 1 | 3 | 3 | 4 | 2 | 1 | 1 | 2 | 1 | 3 | 3 | 1 | 2 | 2 | 3 | 3 | |
| 31 | Sample 58-09 | 1 | – | 2 | 1 | 2 | 1 | 1 | 1 | 3 | 1 | – | 1 | 1 | 1 | 3 | 3 | 4 | 2 | 1 | 1 | 2 | 1 | 3 | 3 | 1 | 1 | 2 | 3 | 3 | |
| 32 | Sample 62-09 | 1 | 1 | – | 1 | 2 | 1 | 1 | 1 | 3 | 2 | – | 1 | 1 | 1 | 3 | 3 | 4 | 2 | 1 | 1 | 2 | 1 | 3 | 3 | 1 | 2 | 2 | 3 | 3 | |
| 33 | MI-07980 | 1 | – | 2 | 1 | 2 | 1 | 1 | 1 | 3 | 1 | – | 1 | 1 | 1 | 3 | 3 | 4 | 2 | 1 | 1 | 2 | 1 | 3 | 3 | 1 | 2 | 2 | 3 | 3 | |
| 34 | MI-071055 | 2 | – | 1 | 2 | 1 | 1 | 1 | 3 | 2 | 1 | 1 | 1 | 1 | 3 | 3 | 4 | 2 | 1 | 1 | 2 | 1 | 3 | 3 | 1 | 2 | 2 | 3 | 3 | ||
| 35 | Slavyanests | 1 | – | 2 | 1 | 2 | 1 | 1 | 1 | 3 | 1 | – | 1 | 1 | 1 | 3 | 3 | 4 | 2 | 1 | 1 | 2 | 1 | 3 | 3 | 1 | 2 | 2 | 3 | 3 | |
| 36 | Fontan | 1 | 1 | – | 1 | 2 | 1 | 1 | 1 | 2 | 2 | – | 1 | 1 | 1 | 3 | 3 | 4 | 2 | 1 | 1 | 2 | 1 | 3 | 3 | 1 | 1 | 2 | 3 | 3 | |
| 37 | Serpantin | 1 | – | 2 | 1 | 2 | 1 | 1 | 1 | 3 | 1 | – | 1 | 1 | 1 | 3 | 3 | 4 | 2 | 1 | 1 | 2 | 1 | 3 | 3 | 1 | 2 | 2 | 3 | 3 | |
| 38 | Sprint | 1 | 1 | – | 1 | 2 | 1 | 1 | 1 | 2 | 3 | 2 | 1 | 1 | 1 | 1 | 3 | 3 | 4 | 2 | 1 | 1 | 2 | 1 | 3 | 3 | 1 | 2 | 2 | 3 | 3 |
| 39 | Snezhinka | 1 | – | 2 | 1 | 2 | 1 | 1 | 1 | 2 | 1 | – | 1 | 1 | 1 | 3 | 3 | 4 | 2 | 1 | 1 | 2 | 1 | 3 | 3 | 1 | 1 | 2 | 3 | 3 | |
| 40 | Lider1 | 1 | 1 | – | 1 | 2 | 1 | 1 | 1 | 3 | 3 | 2 | 1 | 1 | 1 | 1 | 3 | 3 | 4 | 2 | 1 | 1 | 2 | 1 | 3 | 3 | 1 | 2 | 2 | 3 | 3 |
| 41 | Lider 2 | 1 | 1 | – | 1 | 2 | 1 | 1 | 1 | – | 3 | 1 | 1 | 1 | 1 | 1 | 3 | 3 | 4 | 2 | 1 | 1 | 2 | 1 | 3 | 3 | 1 | 2 | 2 | 3 | 3 |
| 42 | Rapan | 1 | – | 2 | 1 | 2 | 1 | 1 | 1 | 3 | 1 | – | 1 | 1 | 1 | 3 | 3 | 4 | 2 | 1 | 1 | 2 | 1 | 3 | 3 | 1 | 2 | 2 | 3 | 3 | |
| 43 | Druzhny | 1 | 1 | – | 1 | 2 | 1 | 1 | 1 | 2 | 3 | 2 | 1 | 1 | 1 | 1 | 3 | 3 | 4 | 2 | 1 | 1 | 2 | 1 | 3 | 3 | 1 | 2 | 2 | 3 | 3 |
| 44 | Regul | 1 | – | 2 | 1 | 2 | 1 | 1 | 1 | 3 | 1 | – | 1 | 1 | 1 | 3 | 3 | 4 | 2 | 1 | 1 | 2 | 1 | 3 | 3 | 1 | 2 | 2 | 3 | 3 | |
| 1 | 2 | 3 | 4 | ||||||||||||||||||||||||||||
| 45 | Izumrud | 1 | 1 | – | 1 | 2 | 1 | 1 | 1 | 2 | 2 | – | 1 | 1 | 1 | 3 | 3 | 4 | 2 | 1 | 1 | 2 | 1 | 3 | 3 | 1 | 1 | 2 | 3 | 3 | |
| 46 | Hasar | 1 | – | 2 | 1 | 2 | 1 | 1 | 1 | 2 | 3 | 1 | – | 1 | 1 | 1 | 3 | 3 | 4 | 2 | 1 | 1 | 2 | 1 | 3 | 3 | 1 | 2 | 2 | 3 | 3 |
| 47 | Yantar | 1 | 1 | – | 1 | 2 | 1 | 1 | 1 | 3 | 2 | 1 | 1 | 1 | 1 | 3 | 3 | 4 | 2 | 1 | 1 | 2 | 1 | 3 | 3 | 1 | 2 | 2 | 3 | 3 | |
| 48 | Atlant | 1 | – | 2 | 1 | 2 | 1 | 1 | 1 | 3 | 1 | – | 1 | 1 | 1 | 3 | 3 | 4 | 2 | 1 | 1 | 2 | 1 | 3 | 3 | 1 | 2 | 2 | 3 | 3 | |
| 49 | Garant | 1 | 1 | – | 1 | 2 | 1 | 1 | 1 | 2 | 3 | 2 | 1 | 1 | 1 | 1 | 3 | 3 | 4 | 2 | 1 | 1 | 2 | 1 | 3 | 3 | 1 | 2 | 2 | 3 | 3 |
| 50 | Madina | 1 | – | 2 | 1 | 2 | 1 | 1 | 1 | 3 | 1 | – | 1 | 1 | 1 | 3 | 3 | 4 | 2 | 1 | 1 | 2 | 1 | 3 | 3 | 1 | 2 | 2 | 3 | 3 | |
| 51 | Flagman | 1 | 1 | – | 1 | 2 | 1 | 1 | 1 | 3 | 2 | – | 1 | 1 | 1 | 3 | 3 | 4 | 2 | 1 | 1 | 2 | 1 | 3 | 3 | 1 | 2 | 2 | 3 | 3 | |
| Madina 1 | 1 | – | 2 | 1 | 2 | 1 | 1 | 1 | 3 | 3 | 1 | 1 | 1 | 1 | 1 | 3 | 3 | 4 | 2 | 1 | 1 | 2 | 1 | 3 | 3 | 1 | 2 | 2 | 3 | 3 | |
| Madina 2 | 1 | 1 | – | 1 | 2 | 1 | 1 | 1 | 3 | 2 | 1 | 1 | 1 | 1 | 3 | 3 | 4 | 2 | 1 | 1 | 2 | 1 | 3 | 3 | 1 | 2 | 2 | 3 | 3 | ||
| 52 | Anait | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 3 | 2 | 1 | 1 | 1 | 1 | 3 | 3 | 4 | 2 | 1 | 1 | 2 | 1 | 3 | 3 | 1 | 2 | 2 | 3 | 3 | ||
| 53 | Altynai | 1 | – | 2 | 1 | 2 | 1 | 1 | 1 | 3 | 1 | – | 1 | 1 | 1 | 3 | 3 | 4 | 2 | 1 | 1 | 2 | 1 | 3 | 3 | 1 | 2 | 2 | 3 | 3 | |
| 54 | Barakat | 1 | 1 | – | 1 | 2 | 1 | 1 | 1 | 2 | 2 | – | 1 | 1 | 1 | 3 | 3 | 4 | 2 | 1 | 1 | 2 | 1 | 3 | 3 | 1 | 1 | 2 | 3 | 3 | |
| 55 | Renar | 1 | – | 1 | 1 | 2 | 1 | 1 | 1 | 3 | 1 | – | 1 | 1 | 1 | 3 | 3 | 4 | 2 | 1 | 1 | 2 | 1 | 3 | 3 | 1 | 2 | 2 | 3 | 3 | |
| 56 | Sharm | 1 | 1 | – | 1 | 2 | 1 | 1 | 1 | 2 | 2 | – | 1 | 1 | 1 | 3 | 3 | 4 | 2 | 1 | 1 | 2 | 1 | 3 | 3 | 1 | 2 | 2 | 3 | 3 | |
| 57 | Novator | 1 | 1 | – | 1 | 2 | 1 | 1 | 1 | 2 | 1 | – | 1 | 1 | 1 | 3 | 3 | 4 | 2 | 1 | 1 | 2 | 1 | 3 | 3 | 1 | 2 | 2 | 3 | 3 | |
| 58 | Aral-202 | 1 | 1 | – | 1 | 2 | 1 | 1 | 1 | 3 | 2 | 1 | 1 | 1 | 1 | 3 | 3 | 4 | 2 | 1 | 1 | 2 | 1 | 3 | 3 | 1 | 2 | 2 | 3 | 3 | |
| 59 | KazRI-5 | 1 | – | 1 | 1 | 2 | 1 | 1 | 1 | 2 | 3 | 1 | 1 | 1 | 1 | 1 | 3 | 3 | 4 | 2 | 1 | 1 | 2 | 1 | 3 | 3 | 1 | 2 | 2 | 3 | 3 |
| 60 | Aru | 1 | – | – | 1 | 2 | 1 | 1 | 1 | 3 | 2 | 1 | 1 | 1 | 1 | 3 | 3 | 4 | 2 | 1 | 1 | 2 | 1 | 3 | 3 | 1 | 1 | 2 | 3 | 3 | |
| 61 | Opytny | 1 | – | 2 | 1 | 2 | 1 | 1 | 1 | 3 | 3 | 2 | 1 | 1 | 1 | 1 | 3 | 3 | 4 | 2 | 1 | 1 | 2 | 1 | 3 | 3 | 1 | 2 | 2 | 3 | 3 |
| 62 | Pakli | 1 | 1 | – | 1 | 2 | 1 | 1 | 1 | 2 | 2 | 2 | – | 1 | 1 | 1 | 3 | 3 | 4 | 2 | 1 | 1 | 2 | 1 | 3 | 3 | 1 | 2 | 2 | 3 | 3 |
Cluster analysis on the similarities and differences in the component composition of reserve proteins showed that rice accessions differ in intensity, and on the presence or absence of individual components in the protein spectrum. All genotypes were distributed in 6 clusters, where cluster №1 was the most numerous and consisted of 18 samples. The genotypes of this cluster were characterized by presence of a protein band with an REM 24 in the spectrum, while the other collectible samples did not have this component. This cluster contains the significant part of Kazakhstan breeding: Bakanasski, Marzhan, Madina, Pakli, KazNIIR 5, Opytny, Altynai. The exceptions were varieties Aral 202 and Aru, which became part of clusters 4 and 5. A significant part of the collection consists of varieties and collectible samples of breeding of the All-Russian Rice Research Institute (Figure 2).
 |
Figure 2: Dendrogram of distribution of genotypes of rice at the similarities and differences in the composition of storage proteins in rice seed
|
These samples showed a considerable variety and were distributed in all six groups based on similarities and differences in the composition of proteins (Table 2).
Table 2: Cluster distribution of rice varieties on the intensity and the presence of components of storage proteins in the electrophoretic spectrum
| Number of clusters | Number of sub clusters | Number of samples | REM | |||||||
| 6 | 7 | 8 | 24 | 26 | 28 | 30 | 80 | |||
| 1 | 5 | 3 | 2 | 1 | – | 2 | 3 | 1-2 | 1 | 1-2 |
| 4 | 1 | 1 | – | 2-3 | 3 | 2 | 1 | 2 | ||
| 3 | 0-1 | 1-2 | 0-1 | 2-3 | 2-3 | 1-2 | 0-1 | 1-2 | ||
| 4 | 1 | – | 1-2 | 2-3 | 3 | 1 | 0-1 | 2 | ||
| 4 | 2 | – | 2 | 2-3 | 3 | 1 | 0-1 | 2 | ||
| 2 | 2 | 10 | 1 | – | 1-2 | – | 3 | 1 | – | 2 |
| 3 | 1- | 2 | 1-2 | – | 2-3 | 1 | – | 1 | ||
| 3 | 2 | 4 | 1 | 1 | – | – | 2 | 2 | – | 1 |
| 4 | 1-2 | 1 | – | – | 3 | 1 | – | 1 | ||
| 4 | 2 | 3 | 2 | 2 | – | – | 2-3 | 1-2 | 0-1 | 2 |
| 3 | 2 | 1-2 | – | – | 3 | 2 | 1 | 2 | ||
| 5 | 4 | 2 | 1 | 1 | – | – | 2 | 1-2 | – | 2 |
| 3 | 1 | 0-1 | – | – | 3 | 2 | – | 2 | ||
| 5 | 1 | 1 | – | – | 3 | 1-2 | 1 | 2 | ||
| 3 | 1-2 | – | 1-2 | – | 3 | 2 | 1 | 1-2 | ||
| 6 | 2 | 4 | – | 1-2 | 0-1 | – | 3 | 1-2 | 0-1 | 1-2 |
| 5 | – | 2 | 1 | – | 3 | 1 | 0-1 | 0-1 | ||
| Note: “-” absence component, “0-1” absence – presence, “1”, “2”, “3” – intensity | ||||||||||
4 samples of cluster №3 showed significant similarity in the spectrum. Samples in the clusters №2 and №3 did not have a component with REM 30, whereas for genotypes of cluster №6 did not have a component with REM 6. Varieties of Russian breeding such as Viola, Kuban 3, Sonata, Kurchanka, Hankaisky 429, Liman, Lebed were grouped in this cluster. The obtained data show a significant similarity of varieties of domestic breeding, and the need to increase the genetic diversity of varieties created through the involvement of genetic sources of foreign origin.
Conclusions
The optimum conditions for extraction and fractionation of rice reserve proteins and a way of components registering were selected in the result of researches.
The most variable area of the protein band of the spectrum is revealed, analyses were done and protein formula for 62 varieties from the rice collection were compiled.
Clustering was carried out based on the similarities and differences in the composition of grain reserve proteins. Substantial similarity of domestic breeding rice varieties for protein characteristics was established.
References
- Sozinov, A.A. Polymorphism of proteins and its importance in genetics and breeding. Moscow. Nauka. 1985; 272 p. [in Russian].
- Konarev, A.V. The using of molecular markers in plant genetic resources. Agricultural Biology. 1998; V.5; pp 3-25. [in Russian].
- Huebner, F.R., Bietz, J.A., Juliano, B.O. Rice cultivar identification by high –performance liquid chromatography of endosperm proteins// Cereal. Chemistry. 1990; V.67; pp. 129-135.
- Tumanyan, N.G., Lotochnikova, T.N. Rice as a raw material for functional purposes rice production. Risovodstvo. 2010; V16; pp. 102- 108. [in Russian].
- Cagampang, G.B., Cruz, L.J., Espiritu, S.G., Santigo, R.G., Juliano, B.O. Studies on the extraction and composition of rice protein. Cereal Chem.1966; V.43; pp. 145-155.
- Jin, W.D., Li, N., Hong, D.L. Genetic diversity of seed storage protein in different ecotype varieties of japonica rice and its application. Rice science. 2006; V. 13; N2; pp.85-92.
- Galani, S., Naz, F., Soomro, F., Jamil, I., Zia-ul-hassan, Azhar A., Ashraf, A. Seed storage protein polymorphism in ten elite rice (Oriza sativa L.) genotypes of Sindh. African Journal of biotechnology. 2011; V. 10; N7; pp. 1106-1111.
- Khan, N., Ali, M., Rabbani, M.A., Masood, M.Sh. Diversity of glutenin alpha subunits in rice varieties from Pakistan. Pac. J. Bot. 2010; V.42; N3; pp. 2051-2057.
- Siddiqui, S.U., Satoh, H., Kumammaru, T. Variation in seed storage proteins of Pakistani rice germplasm // in: Khush G.S., Brar D.S., Hardy B. Advance in rice Genetics. Manila, Philippines: IRRI. 2003; pp. 198-200.
- Santos, K.F.D.N., Silveira, R.D.D., Martin-Didonet, C.C.G., Brondani, C. Storage protein profile and amino acid content in wild rice Oriza glumaepatula//Pesq. Agropec. Bras. 2013; V.48; N1; pp. 66-72.
CrossRef - Lu, Z.M., Wang, H, Shen, Y.J. Polymorphism of endosperm storage protein and application in hybrid rice (Oryza sativa L.) // J. Nanjing Agric.Univ. 2001; V. 24; N2; pp. 6-11.
- Sarker, R., Bose, S. Electrophoresis characterization of rice varieties using single seed (salt soluble) proteins // Theor. Appl. Genet. 1984; V.68; pp. 415-419.
- Prathepha, P., Daipolmak, V., Samappito, S., Baimai, V. An assessment of alkali degradation, Waxy protein and their relation to amylase content in Thai rise cultivars // Science Asia. 2005; V. 31. pp. 69-75.
CrossRef - Hong, D.L., Satob, H., Kumammaru, T., Qu, L.Q., Sadar, U.S. Protein markers of BT type hybrid and its three lines in japonica rice ((Oryza sativa L.) // Chinese J. Rice Sci. 2001; V.16; N3; pp.165-168.
- Laemmli U.K. Clevage of structural proteins during assembly of the head of bacteriophage. T.4. // Nature. 1970; V. 277; N4; pp. 178-189.

This work is licensed under a Creative Commons Attribution 4.0 International License.





