Manuscript accepted on : 25 February 2016
Published online on: 01-03-2016
Using ISSR-Markers for Genetic Diversity of some Representatives of Orchidaceae
Elena Evgenevna Khomann, Irina Yan Gukovna Nam, Vladimir Vasilievitch Zayakin
Federal State-Funded Educational Institution of Higher Professional Education «Bryansk State Academician I.G. Petrovski University». 241036, Bryansk, Bezhitskaya Street, 14
DOI : http://dx.doi.org/10.13005/bbra/2012
ABSTRACT: The present study is directed to determination the genetic diversity of representatives of Orchidaceae family using six inter simple sequence repeat (ISSR) primers. The possibility of using them to assess the level of genetic polymorphism is shown. Monomorphic and polymorphic bands indicating the species and genus determination have been allocated which enables to use them for making genetic formulas and certification of populations. Cluster analyses on the base of UPGMA (unweighted pair group method with arithmetic mean) made it possible to estimate the degree of similarity of the tested species.
KEYWORDS: Genetic diversity; Orchidaceae; ISSR markers
Download this article as:| Copy the following to cite this article: Khomann E. E, Nam I. Y. G, Zayakin V. V. Using ISSR-Markers for Genetic Diversity of some Representatives of Orchidaceae. Biosci Biotech Res Asia 2016;13(1) |
| Copy the following to cite this URL: Khomann E. E, Nam I. Y. G, Zayakin V. V. Using ISSR-Markers for Genetic Diversity of some Representatives of Orchidaceae. Biosci Biotech Res Asia 2016;13(1). Available from: https://www.biotech-asia.org/?p=6868 |
Introduction
The Orchidaceae is one of the largest angiosperm families1,2,3,4. It lists about 750 genera and 20,000-25,000 species5. Most orchid species are found in the tropical regions1. Bryansk region is a unique floristic region, located in an intersection of important natural, botanical and geographical boundaries. This is an important factor of the high level of biodiversity in the region.
The Red Book of the Bryansk region was written in 2004. An electronic, constantly updated database “Rare species of Sudost and Desna interfluve”, containing more than 2,000 entries about location of rare species of the Bryansk region and their phytocenotic confinement was created which became the basis for monitoring the status of their cenopopulations. There are 18 genera and 35 species of orchids in Bryansk region at present6.
Various methods are available for assessing the condition of populations of rare and endangered plant species. One of the main methods is determination of the level of genetic polymorphism. Inter simple sequence repeats (ISSR) were reported to be a reliable method to determine genetic polymorphism, due to the presence of a large number of microsatellite repeats in the genome of plantshis method of molecular analysis was suggested by Zietkiewicz et al. in 1994 and is based on variations between microsatellites found in the regions7,8.
We have chosen the ISSR method to conduct the molecular genetic analysis9.
Materials And Methods
We used DNA of rare and endangered species, collected in the six regions of the Russian Federation: Bryansk, Kaluga, Kursk, Orel, Smolensk and Tula (Epipactis helleborine (L.) Crantz., Epipactis atrorubens (Hoffm.) Besser, Goodyera repens (L.) R. Br., Platanthera bifolia L., Сypripedium calceolus L., Neottia nidus-avis (L.) Rich., Dactylorhiza incarnata (L.) Soo., Dactylorhiza maculate (L.) Soo., Digitalis grandiflora Mill., Lílium martagon L., Iris aphylla L.). DNA of the three tropical species of Bedugul’s Botanical Gardens collection (Indonesia) (Eria multiflora (Blume) Lindl., Dendrobium malbrownii Dockrill and Pholidota gibbosa (Blume) Lindl. ex de Vriese) was also extracted.
DNA was extracted from young leaves and dry leaf tissues at different time using the Cetyl trimethyl ammonium bromide (CTAB) method.
Six primers were selected for each species tested: IS1((GА)7 G(Y)G), IS2((AC)8G), IS3 ((GA)8C), IS4 ((CA)8A), IS5 ((CA)7(R)C) и IS6 ((AG)7(Y)T).
Each 20 μl of the reaction contained 6 μl of genomic DNA, 1 unit Taq DNA polymerase, 2,5 μl dNTP, 90 pmol of each primer, 2 μl PCR reaction buffer and water. The initial denaturation step at 94 °C for 5 min was followed by 40 cycles of amplification: 94 °C for 45 s, annealing for 45 s, and extension at 72 °C for 1,3 min. The final cycle at 72 °C for 7 min. The temperature of fusion was chosen experimentally.
Extraction of DNA and PCR was performed in 1-3 repeats, due to the rarity of the studied plants and the lack of ability to get enough DNA.
The PCR products were electrophoresed in a 2% agarose gels and stained with ethidium bromide. ISSR amplification products were scanned using GelDocXR system (BioRad, USA) and program for processing electrophoregrams Quantity One. To determine the size of the DNA fragments M27 marker (Sibenzime, Moscow) was used.
Data analysis
Molecular genetic certification was carried out on the basis of the approach proposed by Bronnikova for natural populations of representatives of the family Campanulaceae Juss.: Adenophora lilifolia (L.) A.DC.10.
The study included collecting herbarium material, choosing ISSR markers, PCR analysis, processing electrophoregrams and preparation of certification.
Results And Discussion
Analyzed electrophoregrams revealed monomorphic bands which allows to attribute samples to genus or species and polymorphic bands specific to individual populations (Fig. 1).
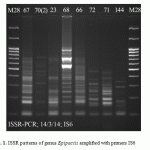 |
Figure 1: ISSR patterns of genus Epipactis amplified with primers IS6 |
Based on this data a type of formulas indicating fragments common to all samples of the studying population and the type of the fragment (g (genus), s (species) and p (polymorphic)) was composed (Table 1). At the beginning of the formula there goes an abbreviation indicating genus in capital letters and small letter indicates species, subscript specifies the type of the fragment, the length of the fragment and the subscript marker used. So the molecular genetic formula for Dactylorhiza maculate with primer IS4, can be written as DTms535IS4. In order to determine whether these fragments belong to the species or taxa of higher rank, the results of molecular genetic analysis of the species belonging to different families were compared
Table 1: Molecular genetic certification of populations
| Population | Number
of samples |
Symbol
of population |
Type of fragment | Molecular genetic formula |
| Cypripedium | 8 | CP | genus (g) | CPg493IS5; CPg460IS5; CPg418IS5; CPg361IS5; CPg300IS5 |
| species (s) | CPms401IS5 | |||
| polymorphic (p) | – | |||
| Dactylorhiza | 12 | DT | genus (g) | DTg190IS1; DTg290IS3; DTg180IS4; DTg337IS6; DTg266IS6; DTg245IS6; DTg218IS6; DTg125IS6 |
| species (s) | DTms535IS4; DTms340IS4; DTms84IS6; | |||
| polymorphic (p) | – | |||
| Epipactis | 12 | EP | genus (g) | EPg60IS1 ;EPg208IS5; EPg100IS6 |
| species (s) | – | |||
| polymorphic (p) | EPhp532IS5; EPhp440IS5; | |||
| Platanthera | 41 | PL | genus (g) | – |
| species (s) | – | |||
| polymorphic (p) | PLbp1176IS1 | |||
| Tropical
orchids |
3 | ERm
DNm PHg |
genus (g) | – |
| species (s) | – | |||
| polymorphic (p) | 64IS4; 300IS6 | |||
| Neottia | 12 | NT | genus (g) | – |
| species (s) | – | |||
| polymorphic (p) | – |
Further, these formulas can be used to compile genetic passports and barcode of populations.
In species of genus Platanthera a total of 2901 DNA fragments were obtained with 6 primers. The highest number of bands (28) was obtained with IS1 primer. Monomorphic bands were not detected. However, with the IS1 primer, 1176 bp band was identified, it is found only in the population of the Bryansk region, Diatkovo area.
Despite the absence of monomorphic bands, there can be identified bands found in most samples. So with primer IS1 a fragment of 200 bp was found in 85%, in 82.5% – of 220 bp, 80% – 280 bp, 57.5% – 330 and 305 bp., with primer IS2 in 55% – 230 bp, with primer IS3 in 87.5% – 200 bp, with primer IS5 in 51.2% – 370 bp and primer IS6 in 92,1% – 160 and 190 bp to 68.4% – 220 bp.
We suppose that the absence of monomorphic bands in studied representatives of the genus Platanthera is connected with high levels of genetic diversity. To obtain more perfect data, it is necessary to collect more plant material from much bigger population, which is difficult due to using herbarium material, rarity of the samples and necessity to save natural populations.
When the study samples of genus Neottia monomorphic bands also weren’t detected. In 54.5% of cases with primer IS1 297 bp and 172 bp bands were identified, in 66.7% – 272 bp and 230 bp, in 58.3% – 320 bp, with primer IS2 in 58,3% – 355 bp., 50% – 500 bp and with primer IS3 in 58,3% – 233 bp.
In order to select extragenic bands and bands of a higher rank DNA from the samples of the families Liliaceae (Lilium martagon), Plantaginaceae (Digitalis grandiflora), Orchidaceae (Goodyera repens) and tropical orchids (Eria multiflora, Dendrobium malbrownii and Pholidota gibbosa) was extracted
In the course of the experiment, we could not find monomorphic bands common to members of all families. However, a fragment 64 bp with IS4 primer and a fragment 300 bp with primer IS6, were found in all samples of tropical orchids. Due to the small number of samples, we cannot assume the relationship with the habitat or location.
Similarities were calculated with Statistica 6.0 computer program and the cluster analysis was performed to develop a dendrogram by unweighted pair group method with arithmetic mean (UPGMA).
The closest accordance data of genetic relationship with location of populations obtained with primers IS4 and IS5 (Fig. 2).
According to this dendrogram samples of Dactylorhiza incarnata: 115 and 116, 114 and 117 are combined in the common clusters that match to their populations of Bryansk region, Sevskii district. Samples Dactylorhiza maculate 107 and 108 are also combined into a common cluster and belong to the population of Kaluga region. On the dendrogram these samples are at the greatest genetic distance from others, and in fact the population is most distant from the others, and most close to it is a sample of Dactylorhiza incarnata 118 (Bryansk region, Diatkovo district), which borders with Kaluga region, which is also shows in the dendrogram.
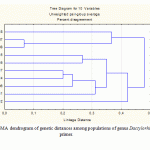 |
Figure 2: UPGMA dendrogram of genetic distances among populations of genus Dactylorhiza with IS4 primer. |
Conclusions
The results of molecular genetic analysis of representatives of Orchidaceae reveal the possibility of using ISSR-primers for analysis of intra- and interspecies differences. The results suggested that the genus Platanthera and Neottia are characterized by the highest level of genetic polymorphism.
Combinations of polymorphic DNA fragments that are specific for the studied genera, species or populations, can be recommend for their genetic certification.
Acknowledgement
This work was supported by the Ministry of Education and Science of the Russian Federation (grant № 41, task № 2014/426).
References
- Haider N., Nabulsi I., Kamary Y. Phylogeny of Orchidaceae species in northwest Syria based on ISSRs. Journal of Plant Biology Research. 2012; 1(2): 36-50.
- López-Roberts M.C., Almeida P.R., Oliveira E.J., van den Berg C. Microsatellite marker development for the threatened orchid Masdevallia solomonii (Orchidaceae). American Journal of Botany. 2012: 66-68.
- Pinheiro L.R., Rabbani A.R.C., da Silva A.V.C., da Silva Le´do A., Pereira K.L.G., Diniz L.E.C. Genetic diversity and population structure in the Brazilian Cattleya labiata (Orchidaceae) using RAPD and ISSR markers. Plant Systematics and Evolution. 2012; 298(10): 1815-1825.
- Dressler R.L., Dodson C.H., 1960. Classification and phylogeny in the Orchidaceae. Annals of the Missouri Botanical Garden. 1960; 47: 25-44.
- Gorovoy P.G., Salokhin A.V., Doudkin R.V., Gavrilenko I.G. Orchidaceae of the Far East: taxonomy, chemical composition, possibilities of protection and use (review). 2010; 13(4): 32–44.
- Evstigneev O.I., Fedotov Iu.P., Panasenko N.N., Velichkin E.M., Kruglikov S.A., Gornov A.V., Radchenko L.A., Bovkunov V.M., Gorokhova A.K. The Red Book of the Bryansk region. Plants. Fungus, Bryansk: Chitai-gorod , 2004; p 267.
- Bhattacharya, S., Bandopadhyay T.K., Ghosh P.D. Efficiency of RAPD and ISSR markers in assessment of molecular diversity in elite germplasms of Cymbopogon winterianus across West Bengal, India. J. Food Agric. 2010; 22 (1): 13-24.
- Efimov P.G. The study of ISSR polimorphizm of Dactylorhiza baltica, fuchsii, D. incarnata (Orchidaceae) from the North-West European Russia. Botanical journal, 2012; 97(6): 751-761.
- Zietkiewicz E., Rafalski A., Labuda D. Genomics, 1994; 20: 176–
- Boronnikova S.V., Nechaeva Y.S. Genetic variation in Ural populations of the rare plant species Adenophora lilifolia (L.) DC. on the basis of analysis of polymorphism of ISSR markers. Bulletin of Perm University, 2012; 1: 41-44.

This work is licensed under a Creative Commons Attribution 4.0 International License.





