Manuscript accepted on :
Published online on: 23-01-2016
Majid Asadi-Ghalehni1, Mohamad Javad Rasaee2, Masoud Javanmardi2, SaeedKhalili2, MohsenMohamadi1, Farnaz Fatemi1 and Esmaeil Sadroddiny*1
1Department of Medical Biotechnology, School of Advanced Technologies in Medicine, Tehran University of Medical Sciences, Tehran, Iran 2Department of Medical Biotechnology, Faculty of Medical Sciences, Tarbiat Modares University, Tehran, I.R. Iran
DOI : http://dx.doi.org/10.13005/bbra/1917
ABSTRACT: The epidermal growth factor receptor (EGFR) belongs to the ErbB family of receptor tyrosine kinases (RTK). EGFR over expressed in different cancers like neck, lung and breast. Various approaches have been used to activate the immune system against EGFR+ cancer such as vaccine therapy. Herein a recombinant fusion peptide composed of EGFR mimotope and L2 domain of EGFR designed. Using in silico studies the potency of the peptide as a vaccine evaluated. This peptide was then cloned into an expression vector and expressed in E. Coli. The ELISA results of the purified peptide against anti EGFR antibodies showed that this peptide could be functional as a vaccine candidate against EGFR-positive tumors.
KEYWORDS: epidermal; receptor; kinases; Evaluation
Download this article as:| Copy the following to cite this article: Asadi-Ghalehni M, Rasaee M. J, Javanmardi M, Khalili S, Mohamadi M, Fatemi F, Sadroddiny E. in Silico and in Vitro Evaluation of A Recombinant Fusion Peptide as A Novel Candidate Vaccine for EGFR-Positive Tumors. Biosci Biotechnol Res Asia 2015;12(3) |
| Copy the following to cite this URL: Asadi-Ghalehni M, Rasaee M. J, Javanmardi M, Khalili S, Mohamadi M, Fatemi F, Sadroddiny E. in Silico and in Vitro Evaluation of A Recombinant Fusion Peptide as A Novel Candidate Vaccine for EGFR-Positive Tumors. Biosci Biotechnol Res Asia 2015;12(3). Available from: https://www.biotech-asia.org/?p=4130 |
Introduction
The receptor for epidermal growth factor (EGFR) is a 170 kDa transmembrane glycoprotein with tyrosine kinase activity, transmiting the mitogenic and inhibitory actions of the EGF family of ligands such as EGF and TGFα (1). Abnormal expression and signaling by EGFR in epithelial cells are associated with tumor initiation and progression (2). This protein has an extracellular ligand-binding domain, a single hydrophobic transmembrane domain and a cytoplasmic tyrosine kinase-containing domain (3). Extracellular domain of the EGFR (ECD-EGFR) is consisted of 4 domains: L1, S1, L2 and S2 (4). It has been revealed that L1 and L2 domains are committed to ligand binding, while L2 domain appears to contribute the most of the ligand binding energy and is more important in EGF binding (5-6). EGFR is involved in some neoplastic processes. It can be over expressed or mutated in many human epithelial tumors such as breast, skin, lung and neck (7).
Various cancer therapy methods, such as application of cancer vaccines, have been developed targeting EGFR. Recently, peptide vaccines have gained more attention as potentially efficient and safe therapeutic modalities against cancers.These kinds of vaccine could be used in immunotherapy of cancers (8).
Bioinformatics has recently emerged as an applicable field to accelerate cancer research. While in silico studies are still ongoing, they play a key role directing the selection of key experiments and the formulation of new testable hypotheses through complete analysis performed by different softwares. For effective vaccine design, epitope mapping of desired protein seems to be necessary (9-10).
Mimotopes, a macromolecule, often a peptide, which mimics the structure of an epitope, and tumor antigen like EGFR, are frequently used in peptide vaccine design (11-13).
Previously, we mapped ICR-62 (an EGFR monoclonal antibody) and reported an EGFR mimotope which was chemically synthesized and its conjugate with BSA was injected into rabbits. It was reported that rabbit IGs against mimotope showed anti EGFR activity (14).
Ramirez et al. showed that using an appropriated adjuvant like FA (freund adjuvant) it would be feasible to stimulate mice immune system to produce anti EGFR antibody against murine extra cellular domain of EGFR (12).
In the present study, we decided to fuse the EGFR mimotope and L2 domain of EGFR to build a novel recombinant protein called EM-L2 (Fig .1). Thereafter, the properties of the EM-L2 were evaluated as a potential vaccine candidate for EGFR+ tumors employing in silico and in vitro methods.
 |
Figure 1: Schematic representation of EM-L2 peptide. EM: EGFR mimotope, L: G4s linker, L2: L2 domain of ECD-EGFR Click here to View figure |
Materials and methods
PDB structure of EGFR and L2 domain –fused to mimotope
Three dimensional structure of Cetuximab (15) and EGFR complex was obtained from Protein Data Bank (PDB) (http://www.pdb.org/) and then Cetuximab (PDB ID: 1YY9) binding site on the EGFR was shown using Pymol software. The sequences of the mimotope, L2 domain of ECD-EGFR and the EM-L2 peptide are listed in Table 1. The sequence of mouse L2 domain (GI:46560582) was retrieved from Entrez protein database, available at NCBI (http://www.ncbi.nlm.nih.gov/) and submitted to m4t server, a fully automated comparative protein structure modeling server (http://www.fiserlab.org/servers/m4t) in order to predict the three dimensional (3D) structure of EM-L2 peptide.
Table 1: the amino acid sequence of the EM-L2 peptide
|
EGFR Mimotpe
L2 Domain of ECD-EGFR
EM-L2 peptide
|
QHYNIVNTQSRV
CTAISGDLHILPVAFKGDSFTRTPPLDPRELEILKTVKEITGFLLIQAWPDNWTDLHAFENLEIIRGRTKQHGQFSLAVVGLNITSLGLRSLKEISDGDVIISGNRNLCYANTINWKKLF
NH2HHHHHHQHYNIVNTQSRVGGGGSCTAISGDLHILPVAFKGDSFTRTPPLDPRELEILKTVKEITGFLLIQAWPDNWTDLHAFENLEIIRGRTKQHGQFSLAVVGLNITSLGLRSLKEISDGDVIISGNRNLCYANTINWKKLFGGGGS-COOH
|
Theoretical physico-chemical properties of the peptides
The theoretical physicochemical properties of the synthetic peptides (such as the ionic status, calculated as the isoelectric point, and the hydrophobicity, measured as the grand average of hydropathicity (GRAVY) index) were analyzed using the ProtParam algorithm (http:// www.expasy.ch/tools/protparam. html). The GRAVY index indicated the hydrophobicity of the peptide and was calculated as the sum of the hydropathy values (Kyte and Doolittle parameters) of the composing amino acids, divided by the number of residues in the sequence. Peptides with positive GRAVY index are hydrophilic whereas peptides with negative GRAVY index are hydrophobic.
B cell epitope prediction
To determine the linear epitopes, the sequence of EM-L2 peptide was submitted to BCPREDS (http://ailab.ist.psu.edu/bcpred/). The sequence similarity index (PD) was computed using SDAP (http://fermi.utmb.edu/SDAP/sdap_pdi.html) server in order to identify the potential EGFR epitopes harboring similar physicochemical properties to the EGFR mimotope.
Construction of expression vector encoding EM-L2 peptide in E.coli
The DNA sequence encoding the EM-L2 peptide was chemically synthesized and amplified by PCR using the forward primer with BamH1 recognition site (5’-TATAGGATCCATGCA TCATCATCATCATCATC-3’) and the reverse primer with EcoRI recognition site (5’-ATATGAATTCTTAACCACCACCGAACAG-3’). The amplified DNA double digested by BamHI and EcoRI (Takara) and ligated into pET32a. The recombinant plasmid “pET32a-EM-L2” was confirmed by restriction endonucleases digestion.
Expression of the recombinant peptide
Competent E.coli BL21 (DE3)pLysS cells were transformed with the recombinant plasmid pET32a-EM-L2 construct. A single clone of E. Coli BL21 (DE3)pLysS harboring the plasmid pET32a-EM-L2 was grown overnight at 37oC in 5 ml LB medium supplemented with 100 mg ampicillin l-1. For expression, 2 ml culture was inoculated into 200 ml freshly prepared LB medium and supplemented with 100 mg ampicillin l-1. The fusion protein expression was induced by addition of IPTG to 0.5mM after the culture reached an OD600 of 0.4–0.6. After 5 h induction, 1 ml of the culture was centrifuged at 5000 rpmfor 5 minutes. The cell pellet was resuspended in 25 ul SDS-PAGE loading buffer for SDS-PAGE analysis.
Purification of the recombinant peptide
500 ml of the culture was centrifuged at 5000 rpm for 5 minutes. The pellet was resuspended in 100 ml PBS and freeze thawed three times at -70 and 30oC. The cells were lysed by sonication on ice for 10 minutes. Lysates were centrifuged at 10000 rpm for 10 minutes and the pellet was washed 3 times with 200 ml PBS containing 3 M urea/l. After washing, the inclusion bodies were solubilizedin suspension buffer (100 mM NaH2PO4, 10 mM Tris/HCl, 8 M urea, pH 8.0) and incubated for 1 h at RT. The suspension was centrifuged at 10000 rpm for 30 minutes at 4oC to remove any remaining insoluble material. The supernatant was transferred to a clean tube and loaded onto a Ni-Sepharose column pre-equilibrated with suspension buffer. The column was washed with 5 bed volumes of washing buffer (100 mM NaH2PO4, 10 mM Tris/HCl, 20 mM imidazole, 8 M urea, pH 8.0). We eluted the protein with a minimal volume of elution buffer (100 mM NaH2PO4, 10 mM Tris/HCl, 500 mM imidazole, pH 8.0) and collected the elution protein for SDS-PAGE and western blot analysis.
Western blot and ELISA analysis for peptide
The purified recombinant proteins were electrophoresis on a 12% (v/v) SDS-PAGE and transferred onto a nitrocellulose membrane (GE Healthcare). After being washed three times with TBST buffer, the membrane was blocked with 5% (v/v) non-fat dry milk in PBS buffer for 90 minutes. The membrane was washed twice for 10 minutes each time with PBS buffer and incubated with HRP-conjugated anti His tag antibody (diluted 1:3000, v/v) (abcam) for 1 h at RT.The membrane was then washed three times with TBST buffer for 10 minutes each. Finally the membrane was analyzed using the DAB substrate.
Table 2: Biochemical properties of the EM-L2 peptide and the highest score B cell epitope of ECD-EGFR.
|
Formula |
C710H1115N207O204S2 |
| Molecular weight | 16342.43 g/mol
|
| Theoretical pI | 8.05 |
| Aliphatic index | 98.17 |
| Instability index | 47.37 |
| Hydropathicity (GRAVY) | -0.278
|
| Estimated solubility | Poor water solubility |
| The highest score B cell epitope of ECD-EGFR | TRTPPLDPRELE (Score:0.999) |
Evaluation of L2D-Em peptide by ELISA method
Wells were coated with 1 µg purified L2D-Em in 100 µl Na2CO3/NaHCO3 buffer and were incubated at 37oC for 5h. Plates were washed two times with TBST and blocked with 5% (w/v) skim milk in TBST for 1 h at RT. Blocking buffer discarded and 1µgr anti EGFR antibodies (ICR61, ICR62 (16) and rabbit IgGs against mimotope (14)) added into the wellsand were incubated for 1 h at 37oC. Wells were washed 5 times with PBST. 100µl of 1/10000 diluted HRP conjugated secondary antibody in PBS (1:10000, v/v) (Pharmacia) was added to the wells and incubated at 37oC for 1 h. After 5 washes with TBST, 50 µl of TMB ELISA substrate solution (Sigma-Aldrich) was added and incubated for 15 minutes at RT. 50 µl of 2M HCL added to stop the reactions. OD values were determined at 450 nm
Results
The 3D structure of the complex EGFR and cetuximab/Erbitux/IMC-C225
The binding site of the Cetuximab on the EGFR molecule is illustrated to be the L2 domain by Pymol software (Figure 2). The 3D structure of the EM-L2 peptide is successfully predicted by mt4 server (http://www.fiserlab.org/servers/m4t ) and shown by Pymol software (Fig 3).
 |
Figure 2 |
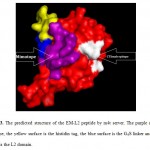 |
Figure 3: The predicted structure of the EM-L2 peptide by m4t server. The purple surface is mimotope, the yellow surface is the histidin tag, the blue surface is the G4S linker and the red surface is the L2 domain. Click here to View figure |
Theoretical physico-chemical properties
Physico-chemical properties of L2D-EM were summarized in table 1. Molecular weight of this peptide is 16342.43 kDa with basic properties (PI: 8.05), while It’s solubility in water is very low (Table 2).
B cell epitope prediction
The linear B cell epitope regions of ECD-EGFR are predicted by BCPREDS server. The highest score of the linear B cell epitopes on ECD-EGFR was located on the L2 domain (Figure 4).
 |
Figure 4: Prediction of some linear B cell epitopes of ECD-EGFR by BCPREDS server. The highest score is for “TRTPPLDPRELE” epitope which is located in the L2 domain of ECD-EGFR. Click here to View figure |
Expression, purification and western blot analysis of recombinant EM-L2 peptide
The bacterial clone harboring the plasmid pET32a–EM-L2 was cultured andinduced by IPTG at the OD600 of 0.4–0.6. The total calculated molecular weight of the peptide and thioredoxin tag was about 28 kDa (Fig 5A). High affinity Ni-NTi resin was used to purify therecombinant protein. Fractions containing the 28 kDa protein (Fig 5B) were dialyzed against urea using PBS (pH 7.4) and concentrated to 500 ul with amicon column (Merck). The yield of recombinant protein was 2.5 mg/l cultures. A Western blot test was carried out with the purified recombinant protein and HRP conjugated anti His tag (abcam) (fig 5C).
 |
Figure 5 Click here to View figure |
Evaluation of L2D-EM peptide
The EM-L2 peptide assessed by ELISA using ICR61, ICR62, rabbit IgGs against the mimotope (14) and irrelevant antibody (anti progesterone) as control performed. Around 50 % OD difference showed that the recombinant peptide can react toward antibodies. The details of results are shown in figure 6.
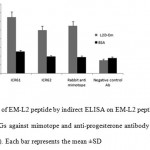 |
Figure 6: Evaluation of EM-L2 peptide by indirect ELISA on EM-L2 peptide. ICR61 and ICR62 antibodies, rabbit IgGs against mimotope and anti-progesterone antibody as irrelevant antibody were used (1 µg/well). Each bar represents the mean ±SD Click here to View figure |
Discussion
The EGFR is a well-defined protein as a tumor associated antigen (TAA) that is expressed in some tumor types. The breadth and depth of technological approaches for making new cancer vaccines has been growing rapidly in recent decades. Peptide vaccination for cancer is one of the most main procedures in this field. Lots of peptides have been introduced as cancer vaccine like EGFR mimotope (14), extra cellular domain of EGFR (12), chemically synthesized peptide (17) and so on. In this study, we considered in silico construction of a new fusion peptide consisted of EGFR mimotope and L2 domain of EGFR. In this regard, the predicted structure interestingly showed that the EGFR mimotope doesn’t cover Cetuximab epitope on L2 domain, so this vital epitope could be exposed property in 3D structure of the EM-L2 peptide too. The linear B- Cell epitope on ECD-EGFR determined. The in silico results showed that this epitope located on the L2 domain of ECD-EGFR. Therefore this domain could be a potential choice for peptide vaccine designing. Baloria et al. mapped the entire HER-2 sequence protein in order to make the entire HER-2 sequence fully immunogenic for DNA vaccine design employing only in silico strategies (18). They claimed their DNA vaccine candidate could produce different antibodies. Although, the antibodies produced against different epitopes of the tumour antigens such as EGFR and HER-2, can exert opposing effects on tumour cell growth (some of them may inhibit tumour growth while others may stimulate it (19).
Herein, in order to have a targeted stimulation of the immune system with fewer complications, a well-established mimotope of EGFR along with the most significant domain of the extra cellular part of EGFR were selected instead of the whole EGFR molecule. Such a peptide could be easily produced in vitro and their function as a vaccine peptide might be much more effective.
Thereafter, the DNA sequence of EM-L2 peptide was synthesized and cloned into an expression vector for evaluation of the hypothesis. E. coli is an excellent host for the production of recombinant peptide antigen because of its efficiency and less costly process (20-21). In this system, the EM-L2 peptide over expressed under the control of strong promoter T7 and the rate of expression was significant. These results indicate that the production of such peptide could be simple and very cheap in E.coli.
Using ELISA method by anti mimotope and anti EGFR antibodies the primary validity of the peptide was proved. The high affinity against the antibodies showed that EM-L2 peptide could be a suitable candidate for EGFR-positive tumors. Moreover, Navari et al. evaluated the EGFR mimotope/ICR62 reaction (14). Our findings confirmed the EM-L2 affinity toward anti EGFR antibodies. However, using such peptide in mice may induce some nonspecific antibodies which their effects on EGFR tumors should be considered.
It is proposed that such a peptide could be considered in phage vaccine and peptide vaccine strategies. We hypothesized that either the displaying of EM-L2 on phage surface or its combination with a suitable adjuvant, and their subsequent injection to EGFR+ mice model could result in reduced tumor growth rate.
In conclusion, it should be noted that there are a lot of strategies for peptide vaccine design. According to our findings it is concluded that fusion of well known peptides and their in silico and in vitro evaluation could result in the construction of novel vaccine peptides.
Acknowledgment
We really appreciate prof. Modjtahedi for his generous gift of ICR61 and ICR62 antibodies.
References
- Salomon DS, Brandt R, Ciardiello F, Normanno N. Epidermal growth factor-related peptides and their receptors in human malignancies. Critical reviews in oncology/hematology. 1995;19(3):183-232.
- Hynes NE, Lane HA. ERBB receptors and cancer: the complexity of targeted inhibitors. Nature Reviews Cancer. 2005;5(5):341-54
- Olayioye MA, Neve RM, Lane HA, Hynes NE. The ErbB signaling network: receptor heterodimerization in development and cancer. The EMBO journal. 2000;19(13):3159-67.
- Jorissen RN, Treutlein HR, Burgess AW, Epa VC, Garrett TP, Ward CW. Characterization of a comparative model of the extracellular domain of the epidermal growth factor receptor. Protein Science. 2000;9(2):310-24.
- Lax I, Fischer R, Ng C, Segre J, Ullrich A, Givol D, et al. Noncontiguous regions in the extracellular domain of EGF receptor define ligand-binding specificity. Cell regulation. 1991;2(5):337-45.
- Lemmon MA, Bu Z, Ladbury JE, Zhou M, Pinchasi D, Lax I, et al. Two EGF molecules contribute additively to stabilization of the EGFR dimer. The EMBO journal. 1997;16(2):281-94.
- Normanno N, De Luca A, Bianco C, Strizzi L, Mancino M, Maiello MR, et al. Epidermal growth factor receptor (EGFR) signaling in cancer. Gene. 2006;366(1):2-16.
- Rosenberg SA, Yang JC, Restifo NP. Cancer immunotherapy: moving beyond current vaccines. Nature medicine. 2004;10(9):909-15.
- De Groot AS. Immunomics: discovering new targets for vaccines and therapeutics. Drug discovery today. 2006;11(5):203-9.
- De Groot AS, Berzofsky JA. From genome to vaccine—new immunoinformatics tools for vaccine design. Methods. 2004;34(4):425-8.
- Riemer AB, Jensen-Jarolim E. Mimotope vaccines: epitope mimics induce anti-cancer antibodies. Immunology letters. 2007;113(1):1-5.
- Ramírez BS, Pestana ES, Hidalgo GG, García TH, Rodríguez RP, Ullrich A, et al. Active antimetastatic immunotherapy in Lewis lung carcinoma with self EGFR extracellular domain protein in VSSP adjuvant. International journal of cancer. 2006;119(9):2190-9.
- Ramírez BS, Alpízar YA, Fernández DRH, Hidalgo GG, Capote AR, Rodríguez RP, et al. Anti-EGFR activation, anti-proliferative and pro-apoptotic effects of polyclonal antibodies induced by EGFR-based cancer vaccine. Vaccine. 2008;26(38):4918-26.
- Navari M, Zare M, Javanmardi M, Asadi-Ghalehni M, Modjtahedi H, Rasaee MJ. Epitope mapping of epidermal growth factor receptor (EGFR) monoclonal antibody and induction of growth-inhibitory polyclonal antibodies by vaccination with EGFR mimotope. Immunopharmacology and immunotoxicology. 2014;36(5):309-15.
- Voigt M, Braig F, Göthel M, Schulte A, Lamszus K, Bokemeyer C, et al. Functional dissection of the epidermal growth factor receptor epitopes targeted by panitumumab and cetuximab. Neoplasia. 2012;14(11):1023-IN3.
- Modjtahedi H, Komurasaki T, Toyoda H, Dean C. Anti-EGFR monoclonal antibodies which act as EGF, TGFa, HB-EGF and BTC antagonists block the binding of epiregulin to EGFR-expressing tumours. International journal of cancer. 1998;75:310-6.
- Kohno M, Horibe T, Haramoto M, Yano Y, Ohara K, Nakajima O, et al. A novel hybrid peptide targeting EGFR-expressing cancers. European journal of cancer. 2011;47(5):773-83.
- Baloria U, Akhoon BA, Gupta SK, Sharma S, Verma V. In silico proteomic characterization of human epidermal growth factor receptor 2 (HER-2) for the mapping of high affinity antigenic determinants against breast cancer. Amino acids. 2012;42(4):1349-60.
- Yip YL, Smith G, Koch J, Dübel S, Ward RL. Identification of epitope regions recognized by tumor inhibitory and stimulatory anti-ErbB-2 monoclonal antibodies: implications for vaccine design. The Journal of Immunology. 2001;166(8):5271-8.
- Retnoningrum DS, Pramesti H, Santika P, Valerius O, Asjarie S, Suciati T. Codon optimization for high level expression of human bone morphogenetic protein–2 in Escherichia coli. Protein expression and purification. 2012;84(2):188-94.
- Sinsuwan S, Yongsawatdigul J, Chumseng S, Yamabhai M. Efficient expression and purification of recombinant glutaminase from Bacillus licheniformis (GlsA) in Escherichia coli. Protein expression and purification. 2012;83(1):52-8.

This work is licensed under a Creative Commons Attribution 4.0 International License.





