Manuscript accepted on : November 09, 2006
Published online on: --
Plagiarism Check: Yes
Jina Tanzadeh¹, Fazel Najafi Yasouri² and Nasser Ghaemi³
1Department of Biology, Islamic Azad Universit,y Branch of Lahijan, Iran.
2Department of Biology, Faculty of Science, Guilan University, Namjo Street, Iran.
3Department of Biotecnology, University College of Science, University of Tehran, Iran.
ABSTRACT: Soil samples were screened for protease- producing bacteria on skimmilk agar medium. Protease-producing bacteria was identified by clear zones of skim hydrolysis around colonies. Thirty-one thermostable protease producing bacteria were isolated from different soil samples (Compost, hot spring and Manures) collected from various parts of north of Iran. These bacteria were examined for various morphological and Biochemical characteristics according of Bergey’s manual of determination bacteriology . the best skim degrader were Bacillus species.
KEYWORDS: Thermophilic Protease; Soil Bacteria; Iran
| Copy the following to cite this article: Tanzadeh J, Yasouri F. N, Ghaemi N. Isolation and Identification of the Thermophilic Protease - Producing Bacteria from Soil of North of Iran. Biosci Biotech Res Asia 2006;3(2a). |
| Copy the following to cite this URL: Tanzadeh J, Yasouri F. N, Ghaemi N. Isolation and Identification of the Thermophilic Protease - Producing Bacteria from Soil of North of Iran. Biosci Biotech Res Asia 2006;3(2a). Available from: https://bit.ly/2XqVRtu |
Introduction
Microbial proteases may be divided into three groups (Keay, 1971) referred to as acid, neutral and alkaline proteases on the basis of their pH optima (Table 1). An alternative method of classification is based on the mechanism of protease action (Hartley ,1960). According to this method there are four main groups:(i)serine proteases;(ii) thiol proteases,(iii) methal proteases and (iv) acid proteases .
Among the various proteases , bacterial proteases are the most significant , compared with animal and fungal proteases. And among bacteria, Bacillus species are specific producers of extracellular protease
Global requirements of thermostable biocatalysts are for greater than those of the mesophiles of which proteases contribute two thirds (Beg et al., 2003).
Thermostable proteases are advantageous in some applications because higher processing temperatures can be employed , resulting in faster reaction rates , increase in solubility of nongaseous reactants and products , and reduced incidence of microbial contamination by mesophilic organisms. Thermophilic bacteria from hot springs produced unique thermostable enzymes (Rao et al., 1998).
In this paper were report isolation and characterization of protease producing Bacillus sp, from soil which might have useful industrial application .
Materials and Methods
Screening ,Isolation and Identification of Microorganisms
Soil samples from different parts of north of Iran (including soils, cattle and birds composts manure , plants composts and hot spring ) were collected. Suspensions were prepared by mixing 10 gram of each samples with 90 ml of sterile distilled water and were shaken vigorously for 2 min. the samples were heated at 60°C for 60 min in water bath. The liquid was serially diluted in sterile saline, and the dilution from 10-1 to 10-7 was poured and spread on to nutrient agar using spread plate and pour plate methods. Nutrient Agar containing : (g/L Agar,15; Yeast extract, 2; Peptone, 5; Sodium chloride, 5.
These plates were sealed in plastic bags to reduced evaporation and incubated at 55°C for 24h. (Chilcott and Wigley, 1993). Some of the colonies that grew on the plates were purified by successive streaking on nutrient agar plates.
Pure culture were maintained on nutrient agar slant stored in a refrigerator and subcultured at 3 month interval, all colonies isolated by this procedure proved to be thermophilic and did not grow in nutrient agar medium at or below 44°C for as long as 7 days.
To screening for protease producing bacteria; the plates were incubated for up to skimmilk agar at 55oC for 24h. A clear zone of skim hydrolysis gave an indication of producing organisms. The colonies with clear zone were picked up isolated for further investigation these were sub-cultured to obtain pure isolated bacteria species using Asilm et al., (2002) methods.
Evaluation of clear zones of each colony was estimated as radius (mm) of the clear zone minus the radius of the colony (Fig. -1).
The diameter of the clear zone represented the protease activity of strains (Table-3). The most active strains was used for former characterization, properties of the isolated bacteria were identified based on cellular and colonial morphology growth condition , gram stain , motility and biochemical tests (Sneeth and Halt, 1986; Watanabe and Hayana, 1993). In the identification of isolated bacteria species , standard taxonomic description from (Sneath, 1986) were used and Bacillus species were identified. (Table 1).
Protease Production
The culture medium used this work for protease production contained(w/v) 0.5% glucose, 0.75%peptone,0.5%MgSO4.7H2O, 0.5% KH2PO4, and 0.01%FeSO4.7H2O maintained at 50°C for 24 to 72 h in a shaking incubator (140rpm). At the end of each fermentation period , the whole fermentation broth was centrifuged at 10.000rpm at 4°C for 15 min and the clear supernatant was used as crude enzyme preparation.
Determination of Protease Activity
Protease activity was determined in triplicate by incubating 500µl of 0.5% azocasein in Tris-HCl buffer with 100µl enzyme solution for 60 minutes at 50oC, reaction was stopped by adding 500µl of 15% Trichloracetic acid (TCA) with shaking. This was left for 15 min and centrifuged at 4°C for 15 min at 3000 rpm. 1 ml of supernatant was added to 1 ml of 1µl NaoH and absorbance was read at 440nm. one unit(u) of protease activity was defined as micromole of substrate converted per minute under standard assay conditions (Table -3).
Results and Discussion
Seventy-five thermophilic bacteria have been isolated from soil samples according to the method (Chilcott and Wigley ,1993). The isolated bacteria were gram- positive and gram-negative and of which fourty of isolated were identified as Bacillus species with central spores and predominantly unswollen cylindrical sporangia.
Thirty of isolated of hydrolyse both starch and skim and twenty of which hydrolysis only starch and twenty-five of them hydrolysis skim, all of isolated were catalase positive, Indole was not formed and acetoin formation was positive. No growth was obtained in nutrient broth containing more than 7% NaCl.
All of them grew in nutrient broth at 30-60°C with an optimal at 50°C, on the basis of these morphological and Biochemical characteristic, isolated strain as (Table -2) were classified in the genus Bacillus. (Sneath,1985) and denominated Bacillus sp. They were B.brevis, (5 strain) B.licheniformis, (4strain), B.subtilis (4 strain), B.polymyxa (2strain), B.mycoides (5 strain), B.cereus (6 strain), B.megaterium (8 strain), B. alvei (3 strain), B.coagulans (3 strain) (Table -2). Watanaba and Hayano (1993) identified B.subtilis, B.Licheniformis, B.cereus and B.megaterium in soil isolations.
Table 1: Properties of microbial extracellular proteases
| Extracellular
proteases |
source
|
pH range | Mol.wt.
(approx.) |
Main uses | ||
| Max activity | Maxactivity | |||||
| Acid proteases | Mainlyfungi | 2-5 | 2-6 | 35.000 | Cheese making Food industry | |
| Neutral proteases | Bacteriaand fungi | 7-8 | 7-9 | 45.000 | Food processing | |
| Alkaline proteases | Bacteriaand fungi | 9-11 | 5-10 | 27.500 | Detergent additive Leather tanning | |
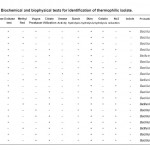 |
Table 1b: Biochemical and biophysical tests for identification of thermophilic isolate. |
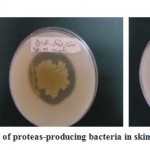 |
Figure 1: Some samples of proteas-producing bacteria in skimmed milk agar plates |
In another investigation (Najafi, 2003) isolated some thermostable alpha-amylase produing bacteria from different soil samples collected from various part of Iran and identified as B.coagulans, B.licheniformis and B.brevis, also Waksman (1991) identified 29 isolated as B.megaterium and 24 isolates as B.subtilis out of 306 soil samples. These agree with the results of this study that Bacillus genera are wide spread among bacteria in soil.
Few publications are devoted to the study of the Bacillus species isolated from various environment. Due to their ubiquity and capability to survive under adverse condition, heterotrophic Bacillus strains and hardly considered to be species of certain habitats (Clus and Berkeley;1986).
The Bacillus genus is an extensive heterogeneous group encompassing 83 validly described species to date. Many species in this taxon one of major clinical importance, such as the B. cereus, B. anthracis, B. thuringiensis, B. mycoides and B. weihenstephanensis), but unfortunately, members of the group shane a great deal of morphological and biochemical similarities (Harrel;L.J, et al., 1995; Priest, et al., 1998). Since the biochemical approach for species identification can be tedious, expensive, and inaccurate, a rapid definitive method is greatly needed. Molecular procedures are increasingly being used for rapid species identification (Jung and Cote; 2002). In contrast, the environmental and nonpathogenic species of this genus exhibit a wide range of physiology DNA base content, and nutritional requirements (Priest, 1993).
Screening for Proteolytic Activity
The proteolytic activity was assayed using skimmilk agar medium and expressed as diameter of clear zone in mm. Strain Jin E,L,P,S exhibited the highest proteolytic activity with a clear zone diameter of 60-65-mm (Figure ,2 , Table 3 ). Table – 3, showed the quantitative assessment of protease productivity by the isolates using fermentation process strains Jin E,L,P,S also produced the highest enzyme activity (806-820 U/ml). Hence it was selected for subsequent investigation .
Table 2a: The morphology and characteristics of some thermophilic isolate
| Isolate | Origin | Cellular | Gram | Spore | motility Growth on | Growth on | Glu | Lac | Suc | Man | |
| code | Morphology Reaction | test | MacConkey | 7%Nacl | |||||||
| Jin A | Plant | Rods | + | _ | + | + | _ | + | _ | _ | _ |
| manure | |||||||||||
| Jin B | Plant | Rods | + | + | + | _ | _ | + | _ | _ | _ |
| compost | |||||||||||
| Jin C | Animal | Rods | + | + | + | _ | _ | + | _ | _ | _ |
| manure | |||||||||||
| Jin D | Animal | Rods | + | + | _ | + | + | + | + | + | + |
| manure | |||||||||||
| Jin E | Water | Rods | + | + | + | + | + | + | + | + | + |
| Spring | |||||||||||
| Jin F | Water | Rods | + | + | _ | _ | + | + | + | + | + |
| spring | |||||||||||
| Jin G | Plant | Rods | + | _ | + | + | _ | + | _ | _ | _ |
| compost | |||||||||||
| Jin H | Animal | Rods | + | + | + | + | + | + | + | + | _ |
| manure | |||||||||||
| Jin J | Water | Rods | + | + | _ | + | + | + | _ | + | + |
| Spring | |||||||||||
| Jin K | Plant | Rods | + | + | _ | + | _ | + | + | + | _ |
| compost | |||||||||||
| Jin L | Plant | Rods | + | + | _ | + | + | + | + | + | + |
| compost | |||||||||||
| Jin M | Animal | Rods | + | + | _ | + | + | + | + | + | + |
| manure | |||||||||||
| Jin N | Animal | Rods | + | + | _ | + | + | + | _ | + | + |
| manure | |||||||||||
| Jin O | Animal | Rods | + | + | _ | + | _ | + | + | + | _ |
| manure | |||||||||||
| Jin P | Water | Rods | + | + | _ | + | + | + | + | + | + |
| Spring | |||||||||||
| Jin Q | Animal | Rods | + | + | _ | + | + | + | + | + | + |
| manure | |||||||||||
| Jin R | Animal | Rods | + | + | _ | + | + | + | _ | + | + |
| manure | |||||||||||
| Jin S | Animal | Rods | + | + | _ | _ | _ | + | + | + | _ |
| manure | |||||||||||
| Jin U | Plant | Rods | + | + | + | + | _ | + | + | + | + |
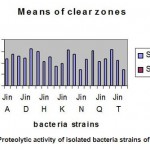 |
Figure 2: Proteolytic activity of isolated bacteria strains of 24h at 55oc |
Table 3: Protease producing ability of isolated strains
| Isolates code | Means of clear zones (mm) | Protease activity (U/ml) |
| JinA | 46 | 650 |
| JinB | 54 | 720 |
| JinC | 52 | 680 |
| JinD | 47 | 600 |
| JinE | 64 | 820 |
| Jin F | 62 | 800 |
| JinG | 41 | 525 |
| JinH | 53 | 750 |
| JinJ | 31 | 350 |
| JinK | 38 | 450 |
| JinL | 62 | 811 |
| JinM | 55 | 780 |
| JinN | 28 | 300 |
| JinO | 45 | 605 |
| JinP | 61 | 810 |
| JinQ | 40 | 400 |
| JinR | 46 | 485 |
| JinS | 63 | 806 |
| JinT | 45 | 420 |
| JinU | 29 | 325 |
Other workers have also isolated thermophilic bacteria in the Thai hot spring (Thailand) , which were reported to produce thermostable proteases (Sookkheo et al., 2000).This might be an indication that the Bacillus species would produce thermostable neutral and alkaline proteases which could find applications in industry and biotechnology .proteases from the Bacillus species are presently being characterized .
References
- Aslim,B.;Saglan , N.;and Beyatli, Y. Determination of some properties of Bacillus isolated from soil. Turk .J.Biol. 26: 41-48 (2002).
- Beg ,KB.;Gupta ,R. Purification of and characterization of an oxidation – stable , Thiol-dependent serin alkaline protease from Bacillus mojavensis. and Microbial Technol., 32: 294-304 (2003).
- Boyer,E.W;and Ingle,M.B. Extracellular alkaline amylase from Bacillus species, Bacteriol.110: 992-1000 (1972).
- Chilcott,C.N.;Wigley ,P.J . Isolation and toxicity of Bacillus thuringiensis from soil and insect habitats in New Zealand, Invert. Pathol. 61, 244-247 (1993).
- Claus D; Berkeley R C W. Genus Bacillus, cohn 18-72. In: Sneath P: H.A, Mair NS; Sharpe ME,Holt, JG (ed) Bergey,s Manual of systematic bacteriology. Baltimore : The Williams and wilkins Co,1105-1139 (1986).
- Dimitrov, P.I.;Kambourova, M.S.; Mandeva.; D.Emanuilova. Isolation and characterization of xylan-degrading, alkali-tolerant thermophiles. FEMS microbial. Lett., 157, 27-30 (1997).
- Gupta , R.;Beg ,QK.;Lovenz , P. Bacterial alkaline protease: Molecular approaches and industrial application. Microbial. Biotechnol. 59: 15-32 (2002).
- Harrel , L.J; Anderson, G.L.,and Wilso, K.H. Genetic variability of anthracis and related species. J.Clin. microbiology. 33: 1947-1950 (1995).
- Joung,D.T.;and Cote, J.C. Evalution of ribosomal RNA gene restriction patterns for the classification of Bacillus species and related genera. Appl.Microbial., 92: 97-108 (2002).
- Najafi,F.;Rasa ,M.;,Sariri ,R. Extraction and Purification of a Thermostable á-amylase from Bacillus coagulans found in Iranian soil samples. J.Chem.Sci., 14, 332-340 (2003).
- Parekh , S.;Vinei , VA.;Stroobel , RJ. Microbiol Biotechnol. 54: 287-301 (2002).
- Pastor ,MD.;Lorda ,GS.;Balatti, A. Protease obtention using Bacillus subtillis 3411 and amaranth seed meal medium at different aeration rates. J. Microbiol., 32: 1-8 (2001).
- Priest ,F.G .;Goodfello.,M and Todd,C. A numerical classification of the genus Bacillus, Gen . Microbial. 134: 1947-1982, (1988).
- Priest, F.G. Systematics and ecology of Bacillus, P369-373.In A.L;Sonenshein,J.A. Hoch and R.losick (ed), Bacillus subtilis and other gram-positiv bacteria:biochemistery, physiology, and molecular genetics:ASM Press,Washington,D.C (1993).
- Rao ,BM.; Tanksale , NA.; Ghatge , SM.; Deshpande , VV. Molecular and Biotechnological Aspects of Microbial Proteases : and Mol. Biol . Rev., 62(3): 591-632 (1998).
- Sneath ,P.H.A. Endospore-forming Grame – positive rods and cocci .In :Sneath, P.H.A, Mair,N.S; Sharpe,M.E and Holt , J.G, Editors, Bergey,s manual of systematic bacteriology., Vol .2, Williams & Willcins , Baltimone, 1104-1139 (1986).
- Stefanova,M.; Emanuilova, E. Characterization of thermostable alpha amylase from Bacillus brevis, EUF .J. , 207: 345-349 (1992).
- Sookkheo , B.,Sinchaikul ,S., Phutrakul , S., Chen , ST. Purification and characterization of the highly thermostable proteases from Bacillus stearothermophilus TLS 33, Protein expression and purification, 20: 142-151 (2000).
- Tonkova , A;Manolov,R;Dobreva, E. Thermostable alpha – amylase from derepressed Bacillus licheniformis in high yield of glucose. Biochem; 28, 539-542 (1993).
- Uzunova,K;Vassileva,A; Mandeva, R; Derekova, A; Ivanova,V, Tonkova, A; Kambourova, M. Production of thermostable innulin-degrading enzyme by thermophilic Bacillus spListy, 94: 972-973 (2000).
- Uguru, G.C.;Akinyanju, J.A. and Sani , A. The use of yam peel for growth of locally isolated Aspergillus niger and α-amylase production.Microbiol .Technol, 21, 48-51 (1997).
- Waksman ,SA. Microbiol Rutgers University. Copyright, print (1961).
- Watanabe,K. ; Hayano ,K. Distribution and identification of proteolytic Bacillus species in paddy field soil under rice cultivation. J.Microbiol., 41, 674-680 (1993).
(Visited 285 times, 1 visits today)

This work is licensed under a Creative Commons Attribution 4.0 International License.





