Manuscript accepted on : 04 October 2017
Published online on: --
Plagiarism Check: Yes
Isolation, Identification and Characterization of Streptomyces Sp. SN-2
Bantanahal Talari Vishwanatha1, Girish Babu K2, Padmashri1, Swathi B Malagi1, Chethan J Dandin2 and Sreenivasa Nayaka1
1Department of Botany, Karnatak University, Dharwad-580003, Karnataka, India.
2Department of Microbiology and Biotechnology, Karnatak University, Dharwad-580003, Karnataka, India.
Corresponding Author E-mail: sreenivasankud@gmail.com
DOI : http://dx.doi.org/10.13005/bbra/2585
ABSTRACT: The present investigation is carried out for the isolation and molecular characterization of actinomycetes from the soil samples from Karnatak University campus, Dharwad, Karnataka, India. Totally four actinomycetes isolated, out of four isolated only one actinomycetes strain (SN-2) showed positive results for antimicrobial activity. SN-2 isolated actinomycetes was further analysed for morphological, physiological, biochemical and 16S ribosomal RNA gene sequencing. The SN-2 strain gene sequence was predicted with secondary structure and restriction sites were identified with the help of Genebee and NEBcutter tools. This investigation clearly indicates that the isolated strain SN-2 as Streptomyces sp.
KEYWORDS: Actinomycetes; Streptomyces; Antimicrobial; 16S rRNA; Cross streak method
Download this article as:| Copy the following to cite this article: Vishwanatha B. T, Babu K. G, Padmashri P, Malagi S. B, Dandin C. J, Nayaka S. Isolation, Identification and Characterization of Streptomyces Sp. SN-2. Biosci Biotech Res Asia 2017;14(4). |
| Copy the following to cite this URL: Vishwanatha B. T, Babu K. G, Padmashri P, Malagi S. B, Dandin C. J, Nayaka S. Isolation, Identification and Characterization of Streptomyces Sp. SN-2. Biosci Biotech Res Asia 2017;14(4). Available from: https://www.biotech-asia.org/?p=28072 |
Introduction
Actinomycetes are unicellular organisms and mode of reproduction is through formation of special types of spores. Actinomycetes are in close relationship with eubactera and they are highly filamentous. It is classified into actinomycetales order, genus actinomycetes it include Streptomyces, Actinomyces, Arthrobacter. Actinomycetes are belonged to gram-positive bacteria and 55% of high Guanine and cytosine (G+C) content present in DNA1. The rare actinomycetes diversity is dominant in aquatic and terrestrial ecosystem and isolated actinomycetes in all most all possible selective methods2,3. Nowadays researchers are more emphasized towards isolation of actinomycetes from different ecological niche. A number of rare actinomycetes were isolated from different regions of soil4,5, marine sources6,7 and hyper saline habitats8. A large number of Streptomyces sp. isolated and screened from soil samples9,10.Therefore the possibilities a novel Streptomyces organisms are isolated from terrestrial habitats have diminished, more than 500 species of actinomycetes are used for the production of secondary metabolites11.
Researchers are searching new genera from soil environments on a regular basis and discovering new metabolites produces never reported earlier. Actinomycete genera identified by cultural and molecular techniques from different marine ecological niches include Streptomyces, Streptosporangium, Nocardiopsis and Microbacterium12. A large number of Streptomyces sp. have been isolated and screened from different habitats in the past several decades13,14. Above 500 species of Streptomyces it produces as 70 to 80% of important secondary metabolites. Actinomycetes are promised source of large bioactive compounds and these are clinical effects and important applications for pharmaceutical and agricultural purpose15,16,17.
Materials and Methods
Collection and Processing of Samples
The soil samples were collected at 2 to 6cm depth from different locations from Karnataka University campus, Dharwad. Totally 10 soil samples are collected using clean stainless steel scoop and a plastic spoon. About 500 g of soil samples were collected in sterile polythene bags. All the collected soil samples were cleaned to remove the stones and debris and air dried for three to four days at room temperature. Cleaned and air dried samples were refilled in the respective polythene bags were stored at 4 °C for further analysis18.
Isolation of Actinomycetes
The soil samples were air dried for 24 h at 40 °C and crushed with mortar and pestle. The isolation of actinomycetes was done by the standard serial dilution method. The standard serial dilution plate culture method was employed to isolate the actinomycetes19. Adequate serial dilution (10-1 to 10-5) were prepared from the enriched soil samples. 0.1 ml of the samples from the respective dilutions were inoculated on starch casein agar medium supplemented with amphotericin B-50 (25µg/ml) and tetracyclin (25µg/ml) and spread using L rod. Finally, incubated at 30 °C for one week, every 24 h we observed the growth of actinomycetes colonies. The actinomycetes isolated colony was observed based on morphological characters like sporulation and pigmentation they were purified as per the method20.
Microscopic Characteristics of Isolated Actinomycetes
Microscopic characters were observed by isolated strains on starch casein agar and glycerol aspergine agar media. The selected isolates were streaked on the surface of the media by streak plate method and plates were incubated at 28 °C for 7 days. Morphological characters such as colony characters, aerial hyphae, substrate mycelium and pigmentation were observed21. The colony colors of the aerial and substrate mycelium were described according to the colors of the RALcode and arrangement of aerial hyphae and spore surface isolates colonies were subsequently observed by phase contrast microscope. For the micro-morphology and external morphology of the SN-2 strain using scanning electron microscopy analysis at national institute for interdisciplinary science and technology (CSIR) Thiruvanantapuram, a sample preparation was determined as per the protocol22.
Cross Streak Method for Antimicrobial Activity
Totally four isolated actinomycetes were tested against bacterial pathogens by using cross streak method23. The pathogenic bacteria used as a test organism for primary screening such as Pseudomonas aeruginosa (P. aeruginosa) (MTCC424), Staphylococcus aureus (S. aureus) (ATTC25923) Escherichia coli (E.coli) (MTCC40), Klebsiella pneumonia (K. pneumonia) (MTCC9238) and Bacillus subtilis (B. substilis) (MTCC121) were obtained from IMTECH, Chandigarh. Activities were assessed using nutrient agar media. Each plate was streaked with each actinomycete isolate at the center of a plate and incubated at 37° for 10 days. These plates were then inoculated with microbial pathogens by streaking at 90° angle to the streak of actinomycetes culture and incubated over night at 37° C. The results were observed based on the inhibition level of the pathogen.
Morphological Characterization of Strain SN-2
The potent active actinomycetes were characterized based on the morphological according to the Bergey’s systematic bacteriology manual24,25. The morphological characteristics were observed in aerial and substrate mycelium and pigmentation in different media such has starch casein agar and glycerol asparagine agar media.
Physiological and Biochemical characteristics
The physiological and biochemical characterization of the active isolated strain SN-7 many tests were performed such as pH, temperature and NaCl tolerance. Specific tests like gram staining, motility, starch, casein, urea, carbohydrate and nitrate reduction test26.
Molecular Characterization of Active Strain SN-2
Amplification of 16s rRNA gene sequence from genomic DNA
The genomic DNA extracted from strain SN-227. The 16S rRNA genes were amplified using primers forward primer GAAGCGCTCACGGCCTA and reverse primer CGGAGTGTCCATGTTCAGGGAACG. The following conditions were used for the PCR amplification. Initial denaturation at 25 cycles of 96 °C for 5 min, followed by 25 cycles of denaturation at 96°C for 30 sec, Hybridization 50°C for 30 sec and final extension 60°C for 1.30 min. The PCR products were electrophoresed on 1% agarose gel with 500 bp DNA ladder as the size reference used. Purified PCR amplicon was sequenced machine (Sequence machine applied biosystems sanger sequencing 3500 series genetic analyzer) and used to interrogate the NCBI database via the BLAST web portal28
Species Identification and 16S rRNA Secondary Structure
The highest similarity of actinomycetes isolated strain SN-2 with the reference species was confirmed using the NCBI BLAST tool (http://www.ncbi.nlm.nih.gov/BLAST). The secondary structure and restriction site was identified by using bioinformatics tools of Genebee and NEBcutter obtained in online software’s (http://WWW.genebee.msu.su/services/rna2_reduced.html, http://tools.neb.com/NEBcutter2) 29.
Results
Isolation of Actinomycetes
Actinomycetes are isolated using different media in starch casein and glycerol aspergine agar media. Isolated actinomycetes are identified according to the Bergey’s manual of systematic bacteriology based on their morphology, sporulation and pigment production in the serially diluted plate 10-1 to 10-5. Totally seven actinomycetes were identified and named as SN-1, SN-2, SN-3, SN-4, SN-5 SN-6 and SN-9, theses strains are tested for gram positive and gram negative bacteria. Out of seven only four isolates shows gram positive bacteria and these four isolates stored on starch casein agar media for further analysis.
Morphological Characterization
Morphological features of isolated colonies of four actinomycetes are described according to the colors of RALcode. The morphological characteristics of four isolates SN-1 as white, SN-2 blue, SN-3 gray and SN-4 as brown color pigmentation was observed.
Antimicrobial Activity
The primary screening of all the four cultures were tested for their inhibitory activity against five bacterial pathogens such as P. aeruginosa, C. bacterium, E. coli, K. pneumonia and B. substalis by streak culture technique. Out of four isolates SN-1, SN-3 and SN-9 showed no inhibition growth from all the tested pathogenic bacteria. Among them, SN-2 showed maximum inhibition growth activity of all the pathogenic bacteria and scanning electron micrograph (SEM) picture shows aerial mycelium of SN-2 strain in Fig 1 and Table 1.
 |
Figure 1: SEM image showing isolated strain SN-2.
|
Table 1: Preliminary screening of antimicrobial activity by cross streak method
|
Actinomycetes strains |
Tested microbes | ||||
| Pseudomonas aeruginosa. | Coryne bacterium | E.coli | Klebsilla pneumonia. | Bacillus substalis. | |
| SN-1 | + | – | – | – | – |
| SN-2 | + | + | + | + | + |
| SN-3 | – | + | – | – | – |
| SN-9 | + | – | + | – | – |
+: Positive inhibition -: No inhibition
Physiological and Biochemical Characterization
The physiological and biochemical test results are summarized Table 2. The Physiological characterization of pH ranges studied from 6.0 to 8.0. The SN-2 showed the optimum growth at pH 7.5. The different temperatures were analysis at 20°C to 40°C. The temperature at 35°C. The different biochemical tests were performed, such as NaCl, starch, casein, urea, gelatine, carbohydrate it shows the positive results and nitrate reduction it shows the negative results of isolated strain SN-2.
Table 2: Physiological and biochemical characterization of potent strain SN-2 isolate
| Test | SN2 |
| Growth of pH | |
| pH6.0 | – |
| pH6.5 | – |
| pH7.0 | + |
| pH7.5 | ++ |
| pH8.0 | + |
| Growth at Temperature | |
| 200C | – |
| 250C | – |
| 300C | + |
| 350C | ++ |
| 400C | + |
| NaCl tolerance | + |
| Hydrolysis of: | |
| Starch | ++ |
| Casein | ++ |
| Urea | ++ |
| Gelatin | ++ |
| Carbohydrate | ++ |
| Nitrate reduction | + |
-: No growth; +: Normal growth; ++: Optimum growth
Molecular Identification of Potential Isolates by 16S rRNA gene Sequencing
The strain SN-2 isolated genomic DNA according to the bacterial genomic DNA isolation kit. The DNA was confirmed with 1% agarose gel and purified. PCR reaction mixture was set up to amplify the 16S rRNA gene. The amplified product was sequenced using ABI Sequencing machine (ABI 3500 XL Genetic Analyzer). The sequence obtained was 853 bp in length and was submitted to NCBI database and Genbank accession numbers are given (Accession No KX284895).
Sequencing and Phylogenetic Tree
The sequence initially characterized by BLAST analysis, which hit homologous 16s rRNA gene from various species. Most homologies of the sequence were found with 16S rRNA genes from Streptomyces sp. SN-2. The DNA sequence was aligned and a phylogenetic tree was constructed by using MEGA4 software (Fig 2).
 |
Figure 2: Dendogram indication the phylogenetic relation of the Streptomyces SN-2
|
Secondary Structure and Restriction Site of Streptomyces sp. SN-2
The RNA secondary structure was predicted for the 16S rRNA gene of Streptomyces sp. SN-2 (Fig 3). This prediction showed that the free energy of the structure is -238.8 kcal/mol, the threshold energy is -4.0 with cluster factor, conserved factor 2 and compensated factor 4 and the conservative is 0.8. The prediction of restriction sites in the strain SN-2 showed the restriction sites for various enzymes, such as BsaB l, SnaBl, EcoR l, Agel and Bsal (Fig 4).
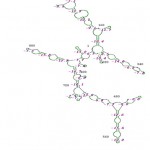 |
Figure 3: Restriction sties on the 16S rRNA gene sequencing of the Streptomyces SN-2
|
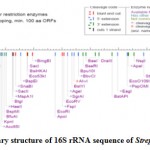 |
Figure 4: Secondary structure of 16S rRNA sequence of Streptomyces sp. SN-2.
|
Discussion
In the present study, we are isolated actinomycetes from the soil samples of the Botanical garden of Karnatak, University, Dharwad. Totally four actinomycetes were isolated and screened for primary screening of antimicrobial activity by cross streak method against different pathogens. The results of the primary screening of antimicrobial activity clearly it indicates that only SN-2 actinomycetes were showed a high activity of inhibition growth for tested pathogenic organisms. These results are compared with the others totally thirty one actinomycetes are isolated from different niche habitats of Sheopur district, Madhya Pradesh and tested their antimicrobial activity of twelve pathogenic microorganisms. These actinomycetes isolates showed antibacterial activity against S. aureus, and less activity against S. dysenteriae30. Similarly actinomycetes are isolated from soil samples of Vengodu (village) in Kanchipuram district, Tamil Nadu, India. Totally thirty five actinomycetes isolated and screened for antibacterial activity by cross streak method and Loyola PBT VAS 10 showed good antibacterial activity against the tested bacteria31.
All the four isolates were grown in starch casein agar and glycerol aspergine agar media, we observed different colors of growth characteristics like aerial, substrate and the pigmentations. Similar findings we observed from actinomycetes are isolated from soil samples in Belgaum, their morphological and cultural characteristics studied of the A-4 mutant showed cellular and aerial growth as well as soluble pigment formation in various ISP media32,33. Actinobacteria have different cultural characteristics in various kinds of culture media, which are important in the classification identification, general with spores, aerial hyphae, with or without color and the soluble pigment, different growth condition on various media as the main characteristics34.
It is always facing to the difficult for identification of actinobacteria. The conventional culture methods are not satisfactory for analysis of actinomycetes. Therefore, 16S rRNA gene sequencing studies are one of the best methods for identification of actinomycetes genera. In the present investigation 16S rRNA amplified and sequenced for isolated strain, SN-2 belongs to the members of Streptomyces sp. Similarly in other studies actinomycetes isolated different niche and identified from 16S rRNA gene sequencing35,36. The RNA secondary structure and restriction sites were predicted by using Genebee and NEBcutter online tools for the 16S rRNA gene of isolated Streptomyces sp. SN-2. Many researchers have been reported using these tools for prediction of secondary structure and restriction sites37,38.
Conclusion
Actinobacteria group of gram positive bacteria and highly G-C content. Actinobacteria are produce variety of antibiotics. Soil has most important habitat for isolation of actinomycetes sp. Actinomycetes are difficult to identification up to species and genus level. The present investigation analysis for 16S rRNA gene sequencing, secondary structure and restriction site analysis. It is concluded that the identified strain SN-2 has Streptomyces sp. and also a good antibiotic producing organism.
Acknowledgement
This research supported by a financial grant from the UGC-SAP-DSA-I at P.G. Department of Botany, Karnatak University, Dharwad, Karnataka-India.
References
- Osada, S., Sutton, A., Muster, N., Brown, C.E., Yates, J.R., Sternglanz, R., and Workman, J.L. The yeast SAS protein complex contains a MYST type putative acetyl transferase and functions with chromatin assembly factor ASF1. Genes and Development., 2001; 15:3155-3168.
CrossRef - Tiwari, K., and Gupta, R.K. Rare actinomycetes: a potential storehouse for novel antibiotics. Critical reviews in biotechnology., 2012; 32(2):108-132.
CrossRef - Tiwari, K., and Gupta, R.K. Diversity and isolation of rare actinomycetes an overview. Critical reviews in biotechnology., 2013; 39(3):256-94.
CrossRef - Li, J., Zhao, G.Z., Huang, H.Y., Zhu, W.Y., Lee, J.C., and Kim C.J. Pseudonocardia rhizophila sp. nov., a novel actinomycete isolated from a rhizosphere soil. Antonie van Leeuwenhoek., 2010; 98(1):77-83.
CrossRef - Ara, I., Bakir, M.A., Hozzein, W.N., and Kudo, T. Population morphological and chemotaxonomical characterization of diverse rare actinomycetes in the mangrove and medicinal plant rhizosphere. African journal of microbial research., 2013; 7(16):1480-1488.
- Radhakrishnan, M., Balagurunathan, R., Selvakumar, N., Doble, M., and Kumar, V. Bioprospecting of marine derived actinomycetes with special reference to antimycobacterial activity. Indian journal of Geomarine science., 2011; 40(3):407-410.
- Goodfellow, M., Stach, J.E., Brown, R., Bonda, A.N., Jones, A.L., and Mexson, J. Verrucosispora marin sp. nov., a novel deep sea actinomycete isolated from a marine sediment which produces abyssomicins. Antonie Van Leeuwenhoek., 2012; 101(1):185-193.
CrossRef - Jose, P.A., and Jebakumar, S.R.D. Phylogenetic appraisal of antagonistic, slow growing actinomycetes isolated from hypersaline inland solar salterns at sambhar salt lake. Frontiers in microbiology., 2013; 4(195):1-9.
CrossRef - Williams, S.T., Goodfellow, M., and Alderson, G. Genus Streptomyces Waksman and Henrici., 1943, 339AL. In: Williams S.T., Sharpe, M.E. and Holt, J.G., Eds., Bergey’s Manual of Systematic Bacteriology., 4, Williams and Wilkins, Baltimore., 1989; 2452-2492.
- Watve, M.G., Tickoo, R., Jog, M.M., and Bhole, B.D. How many antibiotics are produced by the genus Streptomyces. Archives of Microbiology., 2001; 176:386-390.
CrossRef - Malanicheva, I.A., Kozmian, L.I., Dudnik, I.U.V., Stromilova, L.I., and Novozhenov, M.I.U. Protoplast fusion in Streptomyces fradiae strains producing neomycin and tylosin. Antibiot Khimioter., 1992; 37:3-7.
- Ward, A.C., and Bora, N. Diversity and biogeography of marine actinobacteria. Current openion in microbiology., 2006; 9:279-286.
- Hopwood, D.A. How do antibiotic producing bacteria ensure their self resistance before antibiotic biosynthesis incapacitates them? Molecular microbiology., 2007; 63(4):937-940.
CrossRef - Hotam, S.C., Yadav, J., Shrivastava, A.R., Singh, S., Singh, A.K., and Gopalan, N. Antibacterial activity of actinomycetes isolated from different soil samples of Sheopur (A city of central India). Journal of Advanced Pharmaceutical Technology and Research., 2013: 4(2):118-123.
CrossRef - Berdy, J. Bioactive microbial metabolites. The journal of antibiotics., 2005; 58(1):1-26.
CrossRef - Zhao, G.Z., Li, J., Huang, H.Y., Zhu, W.Y., Zhao, L.X., Tang, S.K., Xu, L.H., and Li, W.J. Pseudonocardia artemisiae sp. nov., isolated from surface-sterilized artemisia annua L. International Journal of Systematic and Evolutionary Microbiology., 2011; (61):1061-1065.
CrossRef - Golinska, P., Zucchi, T.D., Silva, L., Dahm, H., and Goodfellow, M. Actinospica durhamensis sp. nov. isolated from a spruce forest soil. Antonie van Leeuwenhoe., 2015; 108:435-442.
CrossRef - Kuster, E., and Locci, R. Studies on peat and peat microorganisms. Archives of Microbiology., 1963; 45(2):188-197.
- Valan Arasu, M., Duraipandiyan, V., Agastian, P., and Ignacimuthu, S. In vitro antimicrobial activity of Streptomyces sp. ERI-3 isolated from Western Ghats rock soil (India). Journal of de Mycologie Medicale., 2009; 19:22-28.
CrossRef - Shirling, E. B., and Gottlieb, D. Methods for characterization of Streptomyces species. International journal of systematic bacteriology., 1966; 16(3):313-340.
CrossRef - Goodfellow., M. Maduromycetes. In Bergey’s manual of systematic bacteriology, 1989; 4:2509-2551. Edited by S. T. Williams, M. E. Sharpe and J. G. Holt. Baltimore: Williams and Wilkins.
- Vijay, S., Nagaraja, M., Sebastian, J., and Ajitkumar, P. Asymmetric cell division in Mycobacterium tuberculosis and its unique features. Archives of Microbiology., 2014; 196:157-168.
CrossRef - Oskay, M. Antifungal and antibacterial compounds from Streptomyces strains. African Journal of Biotechnology., 2009; 8(13):3007-3017.
- Deepika, L., and Kannabiran, K. Biosurfactant and heavy metal resistance activity of Streptomyces sp. isolated from Saltpan soil. British Journal of Pharmacology and Toxicology., 2010; 1:33-39
- Locci, R. Streptomycetes and Related Genera. In Bergey’s Manual of Systematic Bacteriology., 1989. Edited by Williams, S.T. and J.G. Holt, Williams and Wilkins, Baltimore.
- Shirling, E.B., and Gottlieb, D. Methods for characterization of Streptomyces species. International Journal of Systematic Bacteriology., 1966; 16(3):313-340.
CrossRef - Sambrook, J., Fritsch, E.F., and Maniatis,T. Molecular cloning a laboratory manual 2nd ed. Cold Spring Harbor Laboratory, 1989.
- Valan Arasu, M., Duraipandiyan, V., Agastian, P., and Ignacimuthu, S. Antimicrobial activity of Streptomyces sp. ERI-26 recovered from Western Ghats of Tamil Nadu. Journal de Mycologie Medicale., 2008; 18:147-153.
CrossRef - Kaushik, B., Subhro, Banerjee., and Santa, R.J. Diversity of Streptomyces sp. in Eastern Himalayan region-computational RNomics approach to phylogeny. Bioinformation. 2012; 8(12):548-554.
CrossRef - Hotam, S.C., Yadav, J., Shrivastava, A.R., Singh. S., Singh, A.K., and Gopalan, N. Antibacterial activity of actinomycetes isolated from different soil samples of Sheopur (A city of central India). Journal of Advanced Pharmaceutical Technology and Research., 2013; 4(2):118-123.
CrossRef - Pachaiyappan, S.K., John, P.P.R., Veeramuthu, D., and Savarimuthu, I.A. Antimicrobial activity of some actinomycetes from Tamil Nadu, India. Asian Pacific journal of Tropical Biomedicine., 2012; 2(12):936-943.
CrossRef - Nanjwade, B. K., Chandrashekhara, S., Shamarez, A. M., Prakash, S.G., and Fakirappa, V.M. Isolation and morphological characterization of antibiotic producing actinomycetes. Tropical Journal of Pharmaceutical Research., 2010; 9 (3):231-236.
- Taddeii, A., Houwe, G.V., Hediger, F., Kalck, V., Cubizolles, F., Schober, H., and Gasser, S.M. Nuclear pore association confers optimal expression levels for an inducible yeast gene. Nature., 2006; 44:774-778.
CrossRef - Qinyuan, L., Xiu, C., Yi, J., and Chenglin, J. Actinobacteria-Basics and biotechnological applications. Intech press, 2016; pp 87-111.
- Kamil, I., Talha, G., Fadime, O.K., and Elif, C. Molecular identification of different actinomycetes isolated from East Black Sea region plateau soil by 16S rDNA gene sequencing. African Journal of Microbiology Research., 2014; 8(9):878-887.
CrossRef - Thirumurugan, D., and Vijayakumar, R. Characterization and structure elucidation of antibacterial compound of Streptomyces sp. ECR77 isolated from East Coast of India. Current Microbiology., 2015; 70:745-755.
CrossRef - Somvanshi, P., and Seth, P.K. Predicted RNA secondary structures for the conserved regions in dengue virus, Bioinformation., 2010; 3(10):435-439.
CrossRef - Pachaiyappan, S.K., John, P.P.R., Veerumuthum. D., and Savarimuthu. I. Antibacterial activity of some actinomycetes from Tamil Nadu. India Asian pecific journal of Tropical Biomedicine., 2012; 2(12):936-943.

This work is licensed under a Creative Commons Attribution 4.0 International License.





