Manuscript accepted on : 27 September 2017
Published online on: --
Plagiarism Check: Yes
Gene Stacking for Fungal Resistance in Plant Transformation Vector
Sonia Sharma1, Gurtej Singh1, Sadiq Pasha Shaik2, Gagandeep Singh3, Sumangala Bhat4 and Gaurav Sharma1
1Department of Natural Sciences, Sant Baba Bhag Singh University, Jalandhar 144030, India.
2Division of Biotechnology, Indian Institute of Horticulture Research, Hesaraghatta, Bengaluru 560089, India.
3Department of Agriculture, Mata Gujri College, Fatehgarh Sahib 140407, India.
4Department of Biotechnology, University of Agricultural Sciences, Dharwad-580 005, India.
Corresponding Author E-mail: g.sharma7346@gmail.com
DOI : http://dx.doi.org/10.13005/bbra/2563
ABSTRACT: Fungal diseases like early blight, late blight, fusarium wilt cause 30-40 per cent loss in fruit production. Form past decade many transgenic plants had been developed using genes encoding chitinases and glucanases with the objective of imparting fungal disease resistance. Since the genes encoding chitinase and glucanase act synergistically. The study was performed to construct plant transformation vector pRAGS carrying both ech42 and bgn under single T-DNA region. Initially, HindIII site at 5' end of earlier cloned bgn (T. harzianum) was removed using primers during reamplification of the gene. The amplicons were cloned into pTZ57R/T containing T overhangs at Eco321 site and transferred to E. coli DH5a and further to plant transformation vector pBI121 which was named as pRA121. In order to clone another gene (ech42) into pRA121, expression cassette from iHP vector was transferred to pRA121 and named as pRAG121. Further in order to gain XhoI site for cloning ech42 gene into pRAG121, ech42 (pSUM1) was cloned into pYES2/CT, named as pSAG1, ech42 from pSAG1 cloned with KpnI and XhoI in pRAG121 and named as pRAGS121. The vector constructed in the present study can be used to transform important crop plants to have enhanced resistance to fungal diseases.
KEYWORDS: Gene Stacking; Chitinase; Glucanase; Gene pyramiding; kb=Kilobase; bgn= beta glucanase; ech= Endochitinase; PCR= polymerase chain reaction
Download this article as:| Copy the following to cite this article: Sharma S, Singh G, Shaik S. P, Singh G, Bhat S, Sharma G. Gene Stacking for Fungal Resistance in Plant Transformation Vector. Biosci Biotech Res Asia 2017;14(3). |
| Copy the following to cite this URL: Sharma S, Singh G, Shaik S. P, Singh G, Bhat S, Sharma G. Gene Stacking for Fungal Resistance in Plant Transformation Vector. Biosci Biotech Res Asia 2017;14(3). Available from: https://www.biotech-asia.org/?p=27827 |
Introduction
Fungal diseases like early blight, late blight, fusarium wilt cause 30-40 per cent loss in fruit like tomato production (Punja 2006). Form past decade many transgenic plants had been developed using genes encoding chitinases and glucanases with the objective of imparting fungal disease resistance.
In-vivo study showed a collective protective interaction of the co-expressed anti-fungal proteins whereas class I tobacco chitinase and β-1, 3-glucanase acted synergistically. Class II chitinase combined with higher amounts of class I β-1, 3-glucanase showed confined antifungal activity in vitro (Sela-Buurlage et al., 1993). Transgenic wheat plants engineered with chitinase and b-1, 3-glucanase genes showed resistance to scab and powdery mildew (Bliffeld et al., 1999) diseases. Transgenic Brassica napus lines transformed with barley chitinase and b-1, 3-glucanase genes showed improved resistance against Leptosphaeria maculans (Melander et al., 2006).
A number of attempts such as sexual crossing of two different transgenic plants, sequential retransformation (Lapierre et al., 1999), co-transformation with multiple plasmids (Chen et al., 1998 and Hadi et al., 1996) or with single plasmids on which several transgenes are linked (De Gray et al., 2001; Goderis et al., 2002; Akula et al., 2011; Awah et al., 2011; Erika et al., 2013 and Sharad et al., 2015) had been made to introduce multiple genes into plant genomes. These attempts has specific limitations: genetic crosses are time consuming, requirement of different selectable marker genes in sequential retransformation, the efficiency of co-transformation with multiple plasmids is inversely proportional to plasmid number and co-transformation with separate multiple plasmids is a rare event. Therefore it is hard to control their copy number and arrangements among transgenes. In addition, use of biolistics approach for multiple plasmid transformation leads to integration of genes into a few chromosome loci at high copy number, which is not favorable for expression of transgenes (Hadi et al., 1996; Chen et al., 1998; Gelvin et al., 1998 and Maqbool et al., 1999). Hence, co-transformation with linked transgenes in single vectors is a conventional and reliable approach.
Method and Material
Construction of Plant Transformation Vector Carrying Bgn and Ech 42 Gene (Chit 42)
Removal of Restriction Sites in the 5′ End of Bgn Through PCR
The new forward primer was designed to exclude HindIII site at 5′ end of the cloned gene using gene tool software and named as Modglu. Both forward and reverse primers specific to full length bgn were designed earlier (Mala, 2007). This modglu primer was used with the reverse primer to amplify bgn from pSGH2. The sequence of primer used is given below Z-Glu-6 Modglu Forward 5’ ATCAAGATGAAGTACACCATCGTTGCTCCG 3’ Reverse beta 1,6 T. harzianum 5’ GCGCGGCCGCCAATCACTCGTGATTTACC 3’. The purified PCR products of 1.3 kb (50 ng/μl) were ligated to pTZ57R/T vector (2886 bp) as described in InsT/A clone™ PCR product cloning kit (#k1214) from MBI, fermentas USA. These circular plasmids with inserts of 1.3 kb were directly transferred to E. coli DH5a following the protocol mentioned by Sambrook and Russell (2001) with minor modifications. The clone was named as pSG1.
Sub Cloning of Bgn (Restriction Site Modified) Into Plant Transformation Vector
A plant transformation vector pBI121 was used for this purpose. pSG1 and pBI121 is restricted with XbaI and Bam HI. The insert of 1.6 kb size released from pSG1 was eluted and ligated with linearzed pBI121, transferred into E. coli DH5α. The clone was named as pRA121 (carrying CaMV35S: bgn: NOST cassette).
Sub Cloning of Plant Expression Cassette from Ihp Vector to Pra 121
The vector pRA121 and the clones iHP (provided by Dr. Dinesh Kumar, Directorate of Oil seeds Research, Hyderabad, INDIA) were isolated. Digestion of pRA121 and iHP was done with restriction enzymes Hind III. The insert of 1 kb size from iHP (carrying CaMV35S: Catalaseintron: PolyA ) was eluted ligated with linearized pBI121 and transferred into E. coli DH5α. The clone was named as pRAG121 (carrying both CaMV35S: bgn: NOST cassette; CaMV35S: Catalaseintron: PolyA).
Sub Cloning of Endochitanase Genes Into Yeast Expression Vector
In order to have XhoI restriction site from the MCS of pYES2/CT for cloning ech42 gene into pRAG121 previously cloned ech42 gene (Upendra, 2006) was first cloned into yeast expression vector (pYES2/CT). The vector pYES2/CT and the clones pSUM1C (carry ech42) were isolated and restricted with BamHI and KpnI. The insert of 1.6k b size was eluted from pSUM1C. Ligated with linearized pYES2/CT and transferred into E. coli DH5α. The colonies obtained were further streaked on Luria agar with ampicillin (100 μg/ml). The clone was named as pSAG1.
Sub Cloning of ech 42 Gene from pSAG1 vector to pRAG 121
A plant transformation vector pRAG121 carrying both CaMV35S: bgn: NOST cassette; CaMV35S: Catalaseintron: Poly A was used for this purpose. Sequential digestion of pRAG121 and pSAG1 was done with two restriction enzymes Xho I and Kpn I. The insert of 1.6 kb size released from pSAG1 was eluted. Ligated with linerized vector pRAGS121 and transferred into E. coli DH5α.Finaly the catalase intron is replaced by ech 42 and the clone was named as pRAGS121 (carrying CaMV35S: bgn: NOST; CaMV35S:ech42: PolyA cassette)
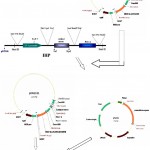 |
Figure 1: Schematic diagram of vector construction carrying both bgn & ech42 gene.
|
Results and Discussion
Construction of Plant Transformation Vector Carrying Both bgn and ech42
Sub Cloning of Restriction Site Modified bgn (gln) from pSGH2
Both the primers were used at 10 pM per μl concentration. The template DNA of pSGH1 gave an amplicon of 1.3 kb was cloned to pTZ57R/T vector. Recombinant cells were selected based on blue/white colony assay. All of these showed the presence of 1.3 kb insert in PCR and restriction analysis (with XbaI and BamHI enzymes). The PCR and restriction analysis of selected clones for sequencing is shown in Fig. 7 and Fig. 10 respectively. The clones were named as pSG1 and the vector map is shown in Fig. 2.
 |
Figure 2: Vector map of pSG1
|
Sub Cloning of bgn (Restriction Site Modified) into Plant Transformation Vector pBI 121
The gene bgn is cloned into pBI121 from pSG1. Plasmid DNA isolated from the clones was confirmed through PCR (Fig. 7) and restriction analysis using XbaI and BamHI enzyme (Fig. 10) and named as pRA121 clones. All of these showed the presence of 1.3 kb insert in PCR and restriction analysis (Fig. 3).
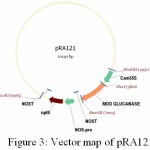 |
Figure 3: Vector map of pRA121
|
Sub Cloning of Cassette from iHP Vector to pRA121
In order to have another plant expression cassette (CaMV35S: Catalase intron: PolyA) to clone ech42 into pRA121, iHP vector was restricted with HindIII to release expression cassate and was cloned in to pRA121. Plasmid DNA isolated from the clones was confirmed through restriction analysis using HindIII enzyme (Fig. 10) and named as pRAG121 (carrying both CaMV35S: bgn: NOST cassette; CaMV35S: Catalase intron: PolyA) clones (Fig. 4). All of these showed the presence of 980 bp insert in PCR and restriction analysis.
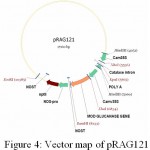 |
Figure 4: Vector map of pRAG121
|
Sub Cloning of the ech42 Into the Yeast Expression Vector
In order to have XhoI restriction site for cloning ech42 into pRAG1 previously cloned ech42 (Upendra, 2006) was first cloned into yeast expression vector (pYES2/CT). Plasmid DNA isolated from the clones was confirmed through PCR (Fig. 8) and restriction analysis using KpnI and BamHI enzyme (Fig. 10) and named as pSAGI clones (Fig. 5). All of these showed the presence of 1.3 kb insert in PCR (Plate 5) and 1.6 kb in restriction analysis (Plate 7).
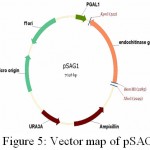 |
Figure 5: Vector map of pSAG1
|
Sub Cloning of ech42 from pSAG1 Vector to pRAG 121
For the construction of a plant transformation vector carrying both bgn and ech42 from Trichoderma, final cloning was performed by inserting ech42 from pSAG1 vector into pRAG121. Plasmid DNA isolated from these clones was confirmed through PCR (Fig. 9) and restriction analysis using XhoI and KpnI enzyme (Fig. 10) and named as pRAGS121 (CaMV35S: bgn: NOST; CaMV35S:ech42: PolyA cassette) clones (Fig. 6). All of these showed the presence of 1.3 kb insert in PCR and 1.6 kb in restriction analysis. Schematic diagram of vector development carrying both ech42 and b-1, 6-glucanase is shown in Fig. 1.
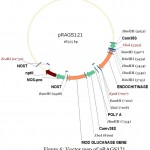 |
Figure 6: Vector map of pRAGS121.
|
Co-transformation with multiple genes in single vectors is a conventional and reliable approach. However, this approach is technically demanding. Our motto of experiment is to construct plant transformation vector (pRAGS121) having both bgn and ech42 genes which act in synergy to degrade fungal cell wall under two separate expression cassette with in a single T-DNA region. On the same T- DNA genes are close to each other. They are tightly linked and will not segregate at higher rate. Further both the genes integrate at the same chromosomal location and will be together in the subsequent generation. Two or more genes, each with its own promoter and terminator, on the same T-DNA region will transfer as a single entity into a plant (i.e. on a single T-DNA for Agrobacterium-mediated transformation). This approach is considered as a special case of co-transformation. Now a day co-transformation with the linking ‘effect’ genes is a preferable strategy that has been used with great advance to stack genes in many GM crops and has gained regulatory approval. Although, a small number of genes (typically two) have been used in this study but many ambitious research projects have used larger linked-gene cassettes. For example, four or five genes (three or four genes related to PHB synthesis, plus a selectable marker gene) have been linked within one T-DNA and introduced into Arabidopsis (Bohmert et al., 2000, 2002) or oilseed rape (Slater et al., 1999). In case if four or more genes needed to be introduced, it is recommended to use several T-DNA cassettes of moderate size (two or three linked ‘effect’ genes) in combination with the co-transformation of the different T-DNAs. Using this strategy, three genes for PHB biosynthesis, plus a selectable marker gene, were introduced on two T-DNAs into Arabidopsis and 16% of the resulting plants produced PHB (Poirier et al., 2000). Similarly, ‘Golden rice’ developed by co-transformation of two T-DNAs, each consisting two linked genes (Ye et al., 2000). Constructs containing as many as 10 foreign DNA fragments had also been created and the Agrobacterium-mediated transfer of 6 linked resistance genes was demonstrated in rice (Lin et al., 2003). Sridevi et al., (2008) used Agrobacterium tumefaciens LBA4404 (pSB1) harbouring the binary vector pNSP3 with rice chitinase and tobacco b-1, 3-glucanase genes (under ubiquitin and CaMV 35S promoter respectively) for transformation of elite indica rice variety Pusa Basmati 1 to enhance sheath blight resistance. Three genes encoding fungal cell wall degrading enzymes (CWDEs), ech42, nag70 and gluc78 from the biocontrol fungus Trichoderma atroviride were inserted into the binary vector pCAMBIA1305.2 singly and in all possible combinations and transformed to rice plants (Liu et al., 2004). Even plasmid construct (pMOG539) carrying four-genes (class I and class II chitinases and /3-1, 3-glucanases) in a single T-DNA under separate CaMV35S promoter were made by assembly of the respective genes next to the neomycin phosphotransferase II (npt ll) gene in the binary plasmid pMOG402 (Jongedijk et al., 1995).
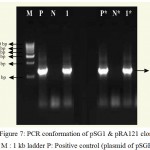 |
Figure 7: PCR conformation of pSG1 & pRA121 clones. M : 1 kb ladder P: Positive control (plasmid of pSGH2),
|
N: Negative control (without plasmid), 1: Amplification of b 1-6, glucanase gene in pSG1,P*: Positive control (plasmid of pSG1), N*: Negative control (without plasmid) and 1*: Amplification of b 1-6, glucanase gene in pRA121.
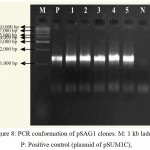 |
Figure 8: PCR conformation of pSAG1 clones. M: 1 kb ladder, P: Positive control (plasmid of pSUM1C),
|
1-5: Amplification of endochitinase gene in pSAG1 andN: Negative control (without plasmid)
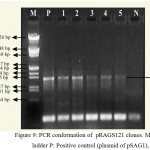 |
Figure 9: PCR conformation of pRAGS121 clones. M: 1 kb ladder P: Positive control (plasmid of pSAG1),
|
1-5: Amplification of endochitinase gene in pRAGS121 and N: Negative control (without plasmid).
This strategy not only overcome the problem associated with segregation but also solves the problem of selection of multiple antibiotic markers in co-transformation with multiple plasmid which leads to poor regeneration due to high selection pressure and also it is tough to have many selectable marker which is not sensitive to crop like rice and tomato. Although the use of large number of selectable markers during multiple plasmid co-transformation is constantly increasing, that would encounter significant hurdles to regulatory approval and public acceptance.
 |
Figure 10: Restriction conformation of pSG1, pRA121, pSAG1& pRAGS121. M1 & M3: l Hind III / EcoRI double digest ladder,
|
1: pSG1 cut with Xba I and Bam HI, 2: pBI121 cut with XbaI and BamHI, 3: pRA121 cut with XbaI and BamHI, 4: iHP cut with HindIII, 5: pRAG121 cut with HindIII, M2: 1 kb ladder 1*: pSAG1 cut with BamHI and KpnI, 2*: pRAGS121 cut with KpnI and XhoI and 3*: pYES2/CT cut with BamHI and KpnI.
One recent Agrobacterium mediated co-transformation experiment with multiple plasmids in Arabidopsis found that most inserts were very complex loci consisting of multiple tandem or inverted T-DNA repeats that often also included the complete binary vector sequence (Stuitje et al., 2003). Such integration patterns leads to transgene silencing in subsequent generations and uphold the regulatory approval of these transgenic crops. Hence, gene stacking with single plasmid in plant transformation vector can give ride of many hurdles coming in way of gene pyramiding in plants and proven to be the most favorable technique to transfer genes which act in synergistic manner.
Acknowledgements
We thank to Department of Biotechnology, New Delhi, India for providing financial support and research scholarships to Masters Students.
Conflict of Interest
There is no conflict of interest among author’s.
References
- Akula N and Dinesh C. A. Enhanced tolerance of transgenic grapevines expressing chitinase and 1,3-glucanase genes to downy mildew. Plant Cell Tiss. Organ Cult. 2011;111:15-28.
- Awah A., Papenbrock S., Jutta B., Fathia H and Jacobsen and Hans-Jörga. Combination of anti fungal genes (Chitinase and Glucanase) to increase the resistance level of transgenic pea (Pisum sativum L.) against fungal diseases. GM Crops. 2011;2(2):104-109.
CrossRef - Bizily S. P., Rugh C. L and Meagher R. B. Phytodetoxification of hazardous organomercurials by genetically engineered plants. Nature biotechnology. 2000;18(2):213.
CrossRef - Bliffeld M., Mundy J., Potrykus I and Fütterer J. Genetic engineering of wheat for increased resistance to powdery mildew disease. Theor. Appl. Gen. 1999;98:1079-1086.
CrossRef - Bohmert K., Balbo I., Kopka J., Mittendorf V., Nawrath C., Poirier Y., Tischendorf G., Trethewey R. N and Willmitzer L. Transgenic Arabidopsis plants can accumulate polyhydroxybutyrate to up to 4% of their fresh weight. Planta. 2000;211:841–845.
CrossRef - Bohmert K., Balbo I., Steinbuchel A., Tischendorf G and Willmitzer L. Constitutive expression of the betaketothiolase gene in transgenic plants. A major obstacle for obtaining polyhydroxybutyrate-producing plants. Plant Physiol. 2002;128:1282–1290.
CrossRef - Chen L., Marmey P., Taylor N. J., Brizard J., Espinoza C., D’Cruz P., Huet H., Zhang S., de Kochko A., Beachy R. N., and Fauquet C. M. Expression and inheritance of multiple transgenes in rice plants. Nature Biotechnol. 1998;16:1060-1064.
CrossRef - DeGray G., Rajasekaran K., Smith F., Sanford J and Daniell H. Expression of an antimicrobial peptide via the chloroplast genome to control phytopathogenic bacteria and fungi. Plant physiology. 2001;127(3):852-862.
CrossRef - Erika L., Alejandrina R., Alejandra M., José O and Eduardo E. Genetic transformation of garlic (Allium sativum L.) with tobacco chitinase and glucanase genes for tolerance to the fungus Sclerotium cepivorum. African J. Biotechnol. 2013;12(22):3482-3492.
- Faize M,, Faize L., Koike N and Ishii H. Acibenzolar-S-methyl-induced resistance to Japanese pear scab is associated with potentiation of multiple defense responses. In. (Ed. GenBank). 2003.
- Faize M., Malnoy M., Dupuis F., Chevalier M., Parisi L and Chevreau E. Chitinases of Trichoderma atroviride induce scab resistance and some metabolic changes in two cultivars of apple. Phytopathol. 2003;93:1496-1504.
CrossRef - Gelvin S. B. Multigene plant transformation more is better. Nature Biotechnology. 1998;16(11):1009-1010.
CrossRef - Goderis I. J., Bolle M. F. D., François I. E., Wouters P. F., Broekaert W. F and Cammue B. P. A set of modular plant transformation vectors allowing flexible insertion of up to six expression units. Plant molecular biology. 2002;50(1):17-27.
CrossRef - Hadi M. Z., McMullen M. D and Finer J. J. Transformation of 12 different plasmids into soybean via particle bombardment. Plant Cell Reports. 1996;15(7):500-505.
CrossRef - Halpin C., Cooke S. E., Barakate A., Amrani A. E and Ryan M. D. Self‐processing 2A‐polyproteins–a system for co‐ordinate expression of multiple proteins in transgenic plants. The Plant Journal. 1999:17(4):453-459.
CrossRef - Jach G., Gornhardt B., Mundy J., Logemann J., Pinsdorf E., Leak R., Schell J and Mass C. Enhanced quantitative resistance against fungal disease by combinational expression of different barley anti-fungal proteins in transgenic tobacco. Plant J. 1995;8:97-109.
CrossRef - Jongedijk E., Tigelaar H., Van Roekel J. S. C., Bres-Vloemans S. A., Dekker I., Van den Elzen P. J. M. Comelissen B. J. C and Melchers L. S. Synergistic activity of chitinases and beta-1, 3- glucanases enhances fungal resistance in transgenic tomato plants. Euphytica. 1995;85:173-180.
CrossRef - Lapierre C., Pollet B., Petit-Conil M., Toval G., Romero J., Pilate G., Leplé J. C., Boerjan W., Ferret V., Nadai V. D., and Jouanin, L., Structural alterations of lignins in transgenic poplars with depressed cinnamyl alcohol dehydrogenase or caffeic acidO-methyltransferase activity have an opposite impact on the efficiency of industrial kraft pulping. Plant Physiology. 1999;119(1):153-164.
CrossRef - Lin L., Liu Y. G., Xu X and Li B. Efficient linking and transfer of multiple genes by a multigene assembly and transformation vector system. Proceedings of the National Academy of Sciences. 2003;100(10):5962-5967.
CrossRef - Liu M. Z., Sun J., Zhu T., Xu G. E.,and Lorito M. H. Enhancing rice resistance to fungal pathogens by transformation with cell wall degrading enzyme genes from Trichoderma atroviride. J. Zhejiang Univ. Sci. 2004;5(2):133-136.
CrossRef - Ma J .K. C., Hiatt A., Hein M., Vine N. D., Wang F., Stabila P., Dolleweerd C. V., Mostov K and Lehner T. Generation and assembly of secretory antibodies in plants. SCIENCE-NEW YORK THEN WASHINGTON. 1995;716-716.
- Mala G. Cloning and expression of endogluconase genes from Trichoderma species in Saccharomyces cerevisiae. M. Sc. (Agri.) Thesis., Univ. Agric. Sci., Dharwad, Karnataka (India). 2007.
- Maqbool S. B and Christou P. Multiple traits of agronomic importance in transgenic indica rice plants: analysis of transgene integration patterns, expression levels and stability. Molecular Breeding. 1999;5(5):471-480.
CrossRef - Melander M., Kamnert I., Happstadius I., Liljeroth E and Bryngelsson T. Stability of transgene integration and expression in subsequent generations of doubled haploid oilseed rape transformed with chitinase and b-1, 3-glucanase genes in a double-gene construct. Plant Cell Rep. 2006;25:942–952.
CrossRef - Poirier Y., Ventre G and Nawrath C. High-frequency linkage of co-expressing T-DNA in transgenic Arabidopsis thaliana transformed by vacuum-infiltration of Agrobacterium tumefaciens. Theor. Appl. Gen. 2000;100:487–493.
CrossRef - Punja Z. K. Recent developments toward achieving fungal disease resistance in transgenic plants. Can.J. Plant Pathol. 2006;28:298-308.
CrossRef - Sambrook J and Russell D. W., Molecular Cloning : A Laboratory Manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, New York. 2001.
- Schickler H and Chet I. Heterologous chitinase gene expression to improve plant defence against phytopathogenic fungi. J. Ind. Mirobiol. Biotechnol. 1997;19:196-201.
CrossRef - Sela-Buurlage M. B., Ponstein A. S., Bres-Vloemans S. A., Melchers L. S., van den Elzen P. J. M and Cornelissen. Only specific tobacco (Nicotiana tabacum) chitinases and b-1, 3-glucanases exhibit antifungal activity. Plant Physiol. 1993;101:857-863.
CrossRef - Sharad U., Sharma G., Moger N., Bhat S and Krishnaraj P. U. Functional validation of plant transformation vector with stacked ech42 and bgn from Trichoderma in tomato for fungal disease resistance. Indian Journal of Genetics and Plant Breeding (The). 2015;75(1):86-92.
CrossRef - Slater S., Mitsky T. A., Houmiel K. L., Hao M., Reiser S. E., Taylor N. B., Tran M., Valentin H. E., Rodriguez D. J., Stone D. A., Padgette S. R., Kishore G and Gruys K. J. Metabolic engineering of Arabidopsis and Brassica for poly(3-hydroxybutyrate-co-3-hydroxyvalerate) copolymer production. Nat. Biotechnol. 1999;17:1011–1016.
CrossRef - Sridevi G., Parameshwari C., Sabapathi N., Raghupathy V And Veluthambi K. Combined expression of chitinase and b-1, 3-glucanase genes in indica rice (Oryza sativa L.) enhances resistant against Rhizoctonia solani. Pl. Sci. 2008;175:283-290. doi:10.1016/j.plantsci. 2008.04.011.
CrossRef - Stuitje A. R., Verbree E. C., Van der Linden K. H., Mietkiewska E. M., Nap J. P and Kneppers T. J. A. Seed-expressed fluorescent proteins as versatile tools for easy (co)transformation and highthroughput functional genomics in Arabidopsis. Plant Biotechnol. J. 2003;1:301–309.
CrossRef - Upendra M. Characterization of native isolates of Trichoderma spp. and cloning of endochitinase gene. M. Sc. (Agri.) Thesis., Univ. Agric. Sci., Dharwad, Karnataka (India). 2006.
- Ye X. D., Al Babili S., Kloti A., Zhang J., Lucca P., Beyer P and Potrykus I. Engineering the provitamin A (beta-carotene) biosynthetic pathway into (carotenoid-free) rice endosperm. Science. 2000;287:303–305.
CrossRef - Zhu Q., Maher E. A., Masoud S., Dixon R. A., Lamb C. J. Enhanced protection against fungal attack by constitutive co-expression of chitinase and glucanase genes in transgenic tobacco. Biotechnol. 1994;12:807-812.
CrossRef

This work is licensed under a Creative Commons Attribution 4.0 International License.





