Manuscript accepted on : 04 April 2016
Published online on: --
Plagiarism Check: Yes
Noeman Ardalan1, Shahab Jamaran2, Farzam Memari3, Kambiz Davari2, Bahareh Rostami2 and Rashid Ramazanzadeh1
1Department of Biology, Urmia Branch, Islamic Azad University, Urmia, Iran.
2Department of Biology, College of science, Sanandaj branch, Islamic Azad University, Sanandaj, Iran.
3Blood Transfusion Research Center, High Institute for Research and Education in Transfusion Medicine, Tehran, Iran.
4Cellular and Molecular Research Center and Microbiology Department, Faculty of Medicine, Kurdistan University of Medical Sciences, Sanandaj- Iran.
Corresponding Author E-mail: atrop_t51@yahoo.com
DOI : http://dx.doi.org/10.13005/bbra/2271
ABSTRACT: The CTX-M producing organism is emerging as a resistance source to oxyimino-cephalosporin like ceftriaxone and ceftazidime. However, the laboratory detection of this gene has not yet been well defined in Sanandaj, Iran. The purpose beyond this study is identifying the prevalence of CTX-M and its risk factors in the community-acquired Klebsiella pneumoniae infections in Sanandaj.In this case-control study, 100 community-acquired Klebsiella pneumoniae strains were used. CTX-M gene was detected using PCR. The probable clonal relation among strains was determined via Rep-PCR. Risk factors associated with CTX-M positive were examined through the univariate logistic regression, Student’s t-test, and Mann-Whitney U test.The polymerase chain reaction test, used to identify CTX-M gene, showed that 37 (37%) Samples, out of 100 were positive. Based on Rep-PCR, 31 genotypes were identified among 37 Samples of CTX-M positive isolates. According to the statistical analysis, the following were the most important independent risk factors in this study: Gestation (p value= 0.036), previous exposure to antibiotic within 3 months (p value= 0.016), having relatives who work in a hospital (p value= 0.001, and distance of under 2 Km from home address to hospital (p value< 0.001). Regarding the increased value of ESBLs-producing strain, it is strongly recommended to use appropriate curative protocols based on the antibiogram. The results of Rep-PCR experiment refute the hypothesis of the clonal spread of one epidemic strain Klebsiella; meaning that not all CTX-M-producing species originate from the same strain and that the gene has extended among various strains. Hence, hospitals and their worker have to have better hygiene, hospital wastes have to be disposed properly, and antibiotics use may help prevent the spread of ESBL resistance only in case of being prescribed by a doctor.
KEYWORDS: CTX-M; Community-acquired; Klebsiellapneumonia; Risk factors; Rep-PCR
Download this article as:| Copy the following to cite this article: Ardalan N, Jamaran S, Memari F, Davari K, Rostami B, Ramazanzadeh R. Risk factors associated with community-acquired CTX-M producing Klebsiellapneumoniae typing by Rep-PCR in Sanandaj, Iran. Biotech Res Asia 2016;13(3). |
| Copy the following to cite this URL: Ardalan N, Jamaran S, Memari F, Davari K, Rostami B, Ramazanzadeh R. Risk factors associated with community-acquired CTX-M producing Klebsiellapneumoniae typing by Rep-PCR in Sanandaj, Iran. Biotech Res Asia 2016;13(3). Available from: https://www.biotech-asia.org/?p=16171 |
Introduction
Since first report of extended-spectrum beta-lactamases (ESBLs) in K. pneumoniae from Germany (1983)1 the CTX-M family of ESBLs is one of the serious danger to global hygiene2to the extent, that in the previous decade it was described as a pandemic.3 CTX-M is the most widespread ESBLs among Enterobacteriaceae that are agents of hospital and community-acquired.2,4 This increased resistance is largely due to the spread of Escherichia coli and Klebsiella pneumoniae containing CTX-M gene.5 Escherichia coli and K. pneumoniae containing CTX-Mare increasingly prevalent in community6 and nosocomial infections.
One of the foundations of infection control is the knowledge obtained from the surveillance studies. According to reports from patients who have CTX-M-producing bacteria, the following risk factors were identified: history and time-span of hospitalization, disease intensity, hospitalization time-span in ICU, catheter and previous exposure to antibiotic.7,8A rapid typing method can help us greatly to examine epidemiology and determine the genetic relations of resistant strains. An appropriate method is proliferation of Repetitive sequences of bacterial DNA through PCR (Semi-automated Repetitive sequence-based polymerase chain reaction) which has these advantages: 1) low expense 2) high discrimination 3) high speed and 4) a reliable device for taxonomy and typing a wide spectrum of Gram negative bacteria and several gram positive bacteria.9,10 This typing method has proved to be able to determine typing more than 100 isolates in a week.11,12
The increased incidence of ESBL attracted our attention to study risk factors in patients who have community-acquired ESBL-producing K. pneumoniae and the genetic relation between strains by the Rep-PCR typing method.
Materials and Methods
Sampling
During the 10-month study period, we collected 100 samples of K. pneumoniae from Outpatients in Sanandaj laboratories and origin of isolation was from urine, blood, wound, sputum and other respiratory samples. Patient information forms were collected in questionnaires. Items in the questionnaire included demographic data (age at the time of study entry and sex), gestation, underlying disease, having relatives (People leaving in the same house) who work in a hospital, distance of home address from hospital and urinary catheter, antibiotic exposure, previous hospitalizations, within the 3 months prior to study entry. The specimens were collected and processed following conventional microbiological procedures for correct management of clinical samples. Finally bacterial strains were storage in LB broth containing glycerol (15%, v/v) for optimal viability.
Confirmation of ESBL production
Confirmatory combination disc diffusion test was carried out for all isolates. A disc ceftazidime (30 μg) alone and ceftazidime plus clavulanic acid (30 μg/10 μg) were placed at a distance of 25 mm on Mueller-Hinton Agar plate inoculated with a bacterial suspension of 0.5 McFarland turbidity standards. After overnight incubation at 37°C, a positive result was interpreted as ≥ 5 mm increase in a zone diameter for ceftazidime in combination with clavulanic acid versus its zone when tested alone.
DNA extraction
Bacterial genomic DNA was extracted via AxyPrep Bacterial Genomic DNA Miniprep Kit (Axygen Biosciences, USA(13 and the extracted DNA was checked by agarose gel electrophoresis.
Detection of CTX-M gene
In this study, we utilized primer CTX-M-F (5′- ACGCTGTTGTTAGGAAGTG-3′), and CTX-M-R (5′- TTGAGGCTGGGTGAAGT-3′).14 This primer proliferates different sorts of CTX-M (CTX-M-1,-3,-12,-15,-22,-30,-32,-33,-38,-52,-57,-58,-60,-61). The size of the final PCR product for this primer is 759 bp.
PCR were performed with initial denaturation (5 minutes at 94 °C) and then 35 cycles of denaturation (45 seconds at 94 °C), annealing (45 seconds at 58 °C), extension (1 minute at 72 °C). Final products were extended by incubation for 7 min at 72°C.PCR product was analyzed by agarose gel electrophoresis (%1.5) and100 bp DNA ladder was used; finally visualized under UV light after staining with ethidium bromide(50 µg/ml).
Rep-PCR
In order to identify the genetic relation between the 100 existing samples we used the typing technique of Rep-PCR for its simplicity and high speed. Rep-PCR reaction was done in the final volume of 25 microliters including 12.5 microliters of PCR Master Mix, 1 microliter of DNA sample, 1 microliter of every primer, and 9.5 microliters of distilled water. The primer pairREP1 (5′-IIIGCGCCGICATCAGGC-3′) and REP2 (5′-ACGTCTTATCAGGCCTAC-3′) was used to amplify putative Rep-like elements in the genomic bacterial DNA.15
Amplification reactions were carried out in Eppendorf thermal cycler, with an initial denaturation (2 minute 95 °C) then 35 cycles of denaturation (1 minute 92 °C), annealing (1 minute 40°C), extension (8 minute 65 °C), with final extension of 8 min at 65 °C.
Rep-PCR products were electrophoresis in 1.5 %agarose gel. The bonds were detected after staining with by ethidium bromide (50 µg/ml), and photographed by ENDURO™ GDS Gel Documentation System.
To do a fingerprinting analysis, a matrix with 1 for the existence of bond and 0 for its absence were made and the relevant dendrogram was drawn by NTSYS v2.02e software using algorithm of the Unweighted Pair-Group Method (UPGMA).
Statistical analysis
In this study, qualitative variables were evaluated with 95% confidence interval using univariate logistic regression. Quantitative variables were analyzed with Student’s t-test, and Mann-Whitney U test. P value≤0.05were considered statistically significant. The statistical analysis software employed was SPSS v20.
Results
In this case-control study, CTX-M gene was detected in 37 out of 100 samples, which is a considerable percent of resistance to beta-lactamases.
Risk factors associated with CTX-M spread
Based on statistical analysis we found risk factors associated with CTX-M spread that is shown in table 1. Moreover, we found statistically significant relation between previous hospitalization within last three months and the presence of CTX-M. We also found out that the longer Mean duration of hospitalization the more effective it will be on the extension of CTX-M-producing strains. A list of underlying disease of the patients is presented in table 2 and it was only in the case of diabetes that we saw a significant relation between the two groups.
In this study, having relative who work in a hospital has been examined as a risk factor; what was not observed in past studies. CTX-M-producing K. pneumoniae were isolated from 11 out of 13 patients either were workers in a hospital or had some relative working there. This indicates the relation between hospital resistance and resistance in the community. Previous exposure to antibiotic within last three months, and home address distance from hospital were other observed risk factors in this study (see table1).
Table 1: Univariate analysis of risk factors associated with increased risk of acquisition of ESBL-producing K. pneumoniae
| Risk factors | Control
(n= 63) |
CTX-M positive
(n= 37) |
OR (95%CI) | P value |
| Gender
Male Female
|
29 34 |
12 25 |
1.77(0.761-4.149) | 0.184 |
| Mean age in years | 40.75 | 49.59 | ———- | 0.006 |
| Gestation | 2 | 6 | 5.903(1.125-30.975) | 0.036 |
| Previous hospitalizations within the 3 months prior to study entry
|
13 | 28 | 11.96(4.547-31.491) | <0.0001 |
| Mean duration of hospitalization in days
|
2 | 5 | ———- | <0.0001 |
| Previous exposure to antibiotic within the 3 months prior to study entry
|
13 | 24 | 2.806(1.208-6.518) | 0.016 |
| Urinary catheter within the 3 months prior to study entry
|
9 | 2 | 0.343(0.070-1.681) | 0.187 |
| Having relatives who work in a hospital
|
2 | 11 | 12.904(2.671-62.336) | 0.001 |
| Distance of home address from hospital
Under 2 Km 2-8 Km Over 8 Km |
13 36 14 |
23 13 1 |
4.319(2.564-15.574) 0.406(0.175-0.940) 0.097(0.012-0.773) |
<0.0001 0.035 0.028 |
Table 2: Multivariate analysis of underlying disease
| Disease | Control
(n= 63) |
CTX-M positive
(n= 37) |
OR | P value | CI (95%) | |
| Lower | Upper | |||||
| Cardiovascular disease | 2 | 2 | 1.399 | 0.764 | 0.156 | 12.519 |
| Pulmonary disease | 9 | 5 | 1.217 | 0.754 | 0.357 | 4.156 |
| Diabetes | 2 | 8 | 7.715 | 0.015 | 1.479 | 40.247 |
| Dialysis | 1 | 3 | 4.773 | 0.203 | 0.431 | 52.828 |
| vesical calculus | 3 | 2 | 0.969 | 0.976 | 0.129 | 7.255 |
| Renal calculus | 8 | 2 | 0.548 | 0.474 | 0.105 | 2.844 |
Rep-PCR
The next stage was trying to determine the genetic relation between the strains. After drawing the dendrogram based on Rep-PCR, we arranged the CTX-M-producing strains: those strains which had 100 percent genetic similarity were put in one pattern and other strains were, each, put in one distinguished pattern, according to which there exist 31 patterns among the 37 CTX-M-producing strains. Thus, 37 strains of CTX-M positive have 31 distinctive genotypes (figure 1).
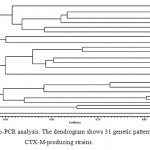 |
Figure 1: Rep-PCR analysis. The dendrogram shows 31 genetic patterns in 37 CTX-M-producing strains. |
Discussion
CTX-M type of ESBL which offers a high resistance to cefotaxime, not to ceftazidime, was described in 199216 and 199317. This family of ESBL is of a class and Ambler classification that is attributed to cefotaximase family (CTX-M). As said before, this type of ESBL was described as a pandemic. We need worldwide studies to find a strategy for preventing the spread of this gene any more. Hence, we identified the risk factors, which help extend the CTX-M-producing strains (table 1 and 2).
In various studies, a relation has been observed between sex and the infection caused by ESBL-producing Enterobacteriaceae18-21. This difference might be due to regional variances in prescribing antibiotics associated with sex (for example, the use of antibiotics to treat the urinary tract infections in women).18inthe present study, sex was not identified as a risk factor. However, Gestation was detected as a factor in extending the infection with strains of CTX-M-producing K. pneumoniae; because urinary tract infections, in which K. pneumoniae play a role, happen more frequently in pregnant women and this by itself leads to increasing the use of antibiotics by such women.
As in our study, previous exposure to antibiotic within the 3 months prior to study entry has been detected as a risk factor in the majority of studies.7,18,19,22-25 By selective pressure, this risk factor may cause eliminating all sensitive strains.
As in other studies, we identified Previous hospitalizations within the 3 months prior to study entry as a risk factor.8,23,25-27 When someone is hospitalized, there are several possibilities for gene transference: 1) that one becomes infected with bacteria containing resistance genes and this resistance may spread among other members of the society; 2) that during hospitalization, the resistance genes stay on one’s skin and ultimately they may be transferred, via transformation mechanism, to those bacteria lacking this gene; 3) this gene goes inside bacteriophages and the hospitalized person carries these viruses outside the hospital and resistance genes get transferred to other bacteria, through transduction mechanism, and the CTX-M gene spreads in the community. We also detected longer Mean duration of hospitalization as a risk factor and this confirms our assumptions.
Urinary catheters, especially when used for a long time, are among the most important factors that help develop urinary tract infection and sometimes bacteremia. In this study, urinary catheter has not been identified as a risk factor. In many studies, as in the case of Arslan et al. (2005), urinary catheter was detected as a risk factor for ciprofloxacin resistance24,and Hayakawa et al. (2013), independent risk factor for CTX-M-producing isolation included presence of a urinary catheter.28 It is possible that urinary catheters be infected with bacteria or the virus containing the gene and in this way CTX-M-producing infection and even other genes increase.
Moreover, having some relatives working in a hospital could be a risk factor for CTX-M extension because the body, especially the hands, of those may be infected with the CTX-M-producing bacteria and this gene can be transferred to other bacteria through the conjugation mechanism; or may be infected with CTX-M gene in which case transformation could be an agent of diffusion; or these genes can placed inside the bacteriophage, the health workers will carry these viruses outside the hospital, the resistance genes may be transfer, through transduction, from hospital bacteria to other bacteria and finally CTX-M gene would spread in the community.
In the case of short distance, home address from hospitals (under 2 Km) was determined as a significant risk factor. Resistance gene from hospital to environment could be transfer via hospital wastes and sewages contain resistance genes, bacteriophages, bacteria containing these genes and so on. Unfortunately, resistance genes will extend outside the hospitals via various gene transfer mechanisms.
The typing technique was necessary in distinguishing the two hypotheses. First: one epidemic Klebsiella strain spread among all patients and so an ancestral strain is responsible for the extension of resistance; this means a relation between all patients. Secondly: CTX-M gene spreads among different Klebsiella strains. We identified 31 different genotypes among 37 samples of CTX-M positive. Thus, the results of this experiment refute the hypothesis of the clonal spread of one epidemic Klebsiella strain; meaning that not all species producing CTX-M originate from the same strain and that the gene has extended among various strains. Accordingly, we cannot contradict the possibility that a plasmid or a transposable element containing CTX-M has caused the spread of the gene among different Klebsiella strains.
Conclusion
In conclusion, we determined the existence of CTX-M gene and we determined associated risk factors in community- acquired infections. Some of these factors were more interesting like that being live in vicinity of hospitals.
Conflict of interest
The authors have no conflict of interest to declare.
Acknowledgement
This is part of Noeman Ardalan M.Sc. thesis. The authors wish to extend their gratitude to the Research Deputy of Kurdistan University of Medical Sciences and Urmia branch Islamic Azad University for financial support.
References
- Bauernfeind A, Schweighart S, Grimm H. A new plasmidic cefotaximase in a clinical isolate of Escherichia coli. 1990;18(5):294-8.
- Bonnet R. Growing group of extended-spectrum β-lactamases: the CTX-M enzymes. Antimicrobial Agents and Chemotherapy. 2004;48(1):1-14.
CrossRef - Cantón R, Coque TM. The CTX-M β-lactamase pandemic. Current Opinion in Microbiology. 2006;9(5):466-75.
CrossRef - Vervoort J, Baraniak A, Gazin M, Sabirova J, Lammens C, Kazma M, et al. Characterization of two new CTX-M-25-group extended-spectrum beta-lactamase variants identified in Escherichia coli isolates from Israel. PLoS One. 2012;7(9):1-7.
CrossRef - Nielsen JB, Skov MN, Jørgensen RL, Heltberg O, Hansen DS, Schønning K. Identification of CTX-M15-, SHV-28-producing Klebsiella pneumoniae ST15 as an epidemic clone in the Copenhagen area using a semi-automated Rep-PCR typing assay. European journal of clinical microbiology& infectious diseases. 2011;30(6):773-8.
CrossRef - Pitout JD, Nordmann P, Laupland KB, Poirel L. Emergence of Enterobacteriaceae producing extended-spectrum β-lactamases (ESBLs) in the community. Journal of Antimicrobial Chemotherapy. 2005;56(1):52-9.
CrossRef - Robino L, Telechea H, Speranza N, Garcia-Fulgueiras V, Cordeiro N, Bado I, et al. Risk factors for the acquisition of extended-spectrum beta-lactamase-producing Enterobacteriaceae in hospitalized children. Journal of infection in developing countries. 2013;7(4):361-4.
CrossRef - Rubio-Perez I, Martin-Perez E, Garcia DD, Calvo ML, Barrera EL. Extended-spectrum beta-lactamase-producing bacteria in a tertiary care hospital in Madrid: epidemiology, risk factors and antimicrobial susceptibility patterns. Emerging health threats journal. 2012;5:1-6.
CrossRef - Olive DM, Bean P. Principles and applications of methods for DNA-based typing of microbial organisms. Journal of Clinical Microbiology. 1999;37(6):1661-9.
- Versalovic J, Schneider M, De Bruijn F, Lupski JR. Genomic fingerprinting of bacteria using repetitive sequence-based polymerase chain reaction. Methods in molecular and cellular biology. 1994;5(1):25-40.
- Pitout JD, Campbell L, Church DL, Wang PW, Guttman DS, Gregson DB. Using a commercial DiversiLab semiautomated repetitive sequence-based PCR typing technique for identification of Escherichia coli clone ST131 producing CTX-M-15. Journal of clinical microbiology. 2009;47(4):1212-5.
CrossRef - Healy M, Huong J, Bittner T, Lising M, Frye S, Raza S, et al. Microbial DNA typing by automated repetitive-sequence-based PCR. Journal of clinical microbiology. 2005;43(1):199-207.
CrossRef - Marcoleta A, Gutiérrez-Cortez S, Maturana D, Monasterio O, Lagos R. Whole-genome sequence of the microcin E492-producing strain Klebsiella pneumoniae RYC492. Genome announcements. 2013;1(1):e00178-13.
CrossRef - Ramazanzadeh R. Etiologic agents and extended-spectrum beta-lactamase production in urinary tract infections in Sanandaj, Iran. Eastern Journal of Medicine. 2010;15:57-62.
- Bou G, Cartelle M, Tomas M, Canle D, Molina F, Moure R, et al. Identification and broad dissemination of the CTX-M-14 beta-lactamase in different Escherichia coli strains in the northwest area of Spain. Journal of clinical microbiology. 2002;40(11):4030-6.
CrossRef - Bauernfeind A, Holley M, Jungwirth R, Mangold P, Röhnisch T, Schweighart S, et al. A new plasmidic cefotaximase from patients infected withSalmonella typhimurium. 1992;20(3):158-63.
- Barthélémy M, Péduzzi J, Bernard H, Tancrède C, Labia R. Close amino acid sequence relationship between the new plasmid-mediated extended-spectrum β-lactamase MEN-1 and chromosomally encoded enzymes ofKlebsiella oxytoca. Biochimica et Biophysica Acta (BBA)-Protein Structure and Molecular Enzymology. 1992;1122(1):15-22.
CrossRef - Ben-Ami R, Rodriguez-Bano J, Arslan H, Pitout JD, Quentin C, Calbo ES, et al. A multinational survey of risk factors for infection with extended-spectrum beta-lactamase-producing enterobacteriaceae in nonhospitalized patients. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America. 2009;49(5):682-90.
CrossRef - Martinez JA, Aguilar J, Almela M, Marco F, Soriano A, Lopez F, et al. Prior use of carbapenems may be a significant risk factor for extended-spectrum beta-lactamase-producing Escherichia coli or Klebsiella spp. in patients with bacteraemia. The Journal of antimicrobial chemotherapy. 2006;58(5):1082-5.
CrossRef - Colodner R, Rock W, Chazan B, Keller N, Guy N, Sakran W, et al. Risk factors for the development of extended-spectrum beta-lactamase-producing bacteria in nonhospitalized patients. European journal of clinical microbiology & infectious diseases : official publication of the European Society of Clinical Microbiology. 2004;23(3):163-7.
CrossRef - Neulier C, Birgand G, Ruppé É, Armand-Lefèvre L, Lolom I, Yazdanpanah Y, et al. Enterobacteriaceae bacteremia: Risk factors for ESBLPE. Médecine et Maladies Infectieuses. 2014;44(1):32-8.
CrossRef - Kaya O, Akcam FZ, Gonen I, Unal O, Ceylan T. Risk factors for bacteremia due to extended-spectrum beta-lactamase-producing Escherichia coli in a Turkish hospital. Journal of infection in developing countries. 2013;7(7):507-12.
CrossRef - Tumbarello M, Spanu T, Sanguinetti M, Citton R, Montuori E, Leone F, et al. Bloodstream infections caused by extended-spectrum-beta-lactamase-producing Klebsiella pneumoniae: risk factors, molecular epidemiology, and clinical outcome. Antimicrobial agents and chemotherapy. 2006;50(2):498-504.
CrossRef - Arslan H, Azap OK, Ergonul O, Timurkaynak F. Risk factors for ciprofloxacin resistance among Escherichia coli strains isolated from community-acquired urinary tract infections in Turkey. The Journal of antimicrobial chemotherapy. 2005;56(5):914-8.
CrossRef - Young BE, Lye DC, Krishnan P, Chan SP, Leo YS. A prospective observational study of the prevalence and risk factors for colonization by antibiotic resistant bacteria in patients at admission to hospital in Singapore. BMC Infectious Diseases. 2014;14(1):1-7.
CrossRef - Calbo E, Romani V, Xercavins M, Gomez L, Vidal CG, Quintana S, et al. Risk factors for community-onset urinary tract infections due to Escherichia coli harbouring extended-spectrum beta-lactamases. The Journal of antimicrobial chemotherapy. 2006;57(4):780-3.
CrossRef - Tham J, Odenholt I, Walder M, Andersson L, Melander E. Risk factors for infections with extended-spectrum beta-lactamase-producing Escherichia coli in a county of Southern Sweden. Infection and drug resistance. 2013;6:93-7.
CrossRef - Hayakawa K, Gattu S, Marchaim D, Bhargava A, Palla M, Alshabani K, et al. Epidemiology and risk factors for isolation of Escherichia coli producing CTX-M-type extended-spectrum beta-lactamase in a large U.S. Medical Center. Antimicrobial agents and chemotherapy. 2013;57:4010-8.
CrossRef

This work is licensed under a Creative Commons Attribution 4.0 International License.





